Bulletin of Botanical Research ›› 2021, Vol. 41 ›› Issue (3): 354-361.doi: 10.7525/j.issn.1673-5102.2021.03.005
• Research report • Previous Articles Next Articles
Yun TENG1, Zong-Bo QIU2( ), Wen-Li WANG2, Xue-Ru ZHANG2
), Wen-Li WANG2, Xue-Ru ZHANG2
Received:2020-04-25
Online:2021-05-20
Published:2021-03-24
Contact:
Zong-Bo QIU
E-mail:qiuzongbo@126.com
About author:TENG Yun(1974—),female,master,lecturer,mainly engaged in the study of development biology in plant.
Supported by:CLC Number:
Yun TENG, Zong-Bo QIU, Wen-Li WANG, Xue-Ru ZHANG. Transcriptome Analysis in Vascular Cambium and Xylem of Paulownia tomentosa[J]. Bulletin of Botanical Research, 2021, 41(3): 354-361.
Add to citation manager EndNote|Ris|BibTeX
URL: https://bbr.nefu.edu.cn/EN/10.7525/j.issn.1673-5102.2021.03.005
Table 1
Summary of transcriptome assembly for P.tomentosa
长度区间 Length interval | 转录本 Transcript | 单基因簇 Unigene |
|---|---|---|
| 200~500 | 77 452 | 68 865 |
| 500~1 000 | 26 725 | 17 742 |
| 1 000~2 000 | 24 155 | 10 848 |
| >2 000 | 17 549 | 6 977 |
| 总数量Total number | 145 881 | 104 432 |
| 总长度Total length | 129 567 733 | 69 112 491 |
| N50长度N50 length | 1 605 | 1 105 |
| 平均长度Mean length | 888 | 662 |
Table 2
Functional annotation of P.tomentosa Unigenes
数据库 Database | 注释Unigenes数 Number of annotated Unigenes | 占注释Unigenes百分数 Percentage of annotated Unigenes (%) |
|---|---|---|
注释到Nr Annotated in Nr | 40 789 | 39.05 |
注释到NT Annotated in NT | 31 675 | 30.33 |
注释到SwissProt Annotated in SwissProt | 30 499 | 29.20 |
注释到GO Annotated in GO | 29 168 | 27.93 |
注释到Pfam Annotated in Pfam | 28 828 | 27.6 |
注释到COG Annotated in COG | 15 539 | 14.87 |
注释到KEGG Annotated in KEGG | 16 316 | 15.62 |
注释到至少一个数据库 Annotated in at least one database | 48 214 | 46.16 |
注释到所有数据库 Annotated in all database | 7 386 | 7.07 |
Unigenes总量 Total Unigenes | 104 432 | 100 |

Fig.1
GO functional classification of unigenes of transcriptome for P.tomentosa vascular cambium and xylem1.Extracellular region;2.Virion part;3.Other organism;4.Membrane part;5.Cell part;6.Synapse part;7.Cell junction;8.Virion;9.Organelle part;10.Extracellular region part;11.Nucleoid;12.Extracellular matrix;13.Other organism part;14.Extracellular matrix component;15.Cell;16.Macromolecular complex;17.Membrane;18.Organelle;19.Synapse;20.Symplast;21.Membrane-enclosed lumen;22.Response to stimulus;23.Cell killing;24.Biological adhesion;25.Developmental process;26.Cellular process;27.Growth;28.Behavior;29.Multi-organism process;30.Cellular component organization or biogenesis;31.Biological regulation;32.Negative regulation of biological process;33.Rhythmic process;34.Reproduction;35.Reproductive process;36.Biological phase;37.Signaling;38.Localization;39.Locomotion;40.Positive regulation of biological process;41.Metabolic process;42.Single-organism process;43.Multicellular organismal process;44.Immune system process;45.Regulation of biological process;46.Nucleic acid binding transcription factor activity;47.Catalytic activity;48.Binding;49.Transporter activity;50.Antioxidant activity;51.Molecular transducer activity;52.Metallochaperone activity;53.Transcription factor activity protein binding;54.Structural activity;55.Molecular function regulator
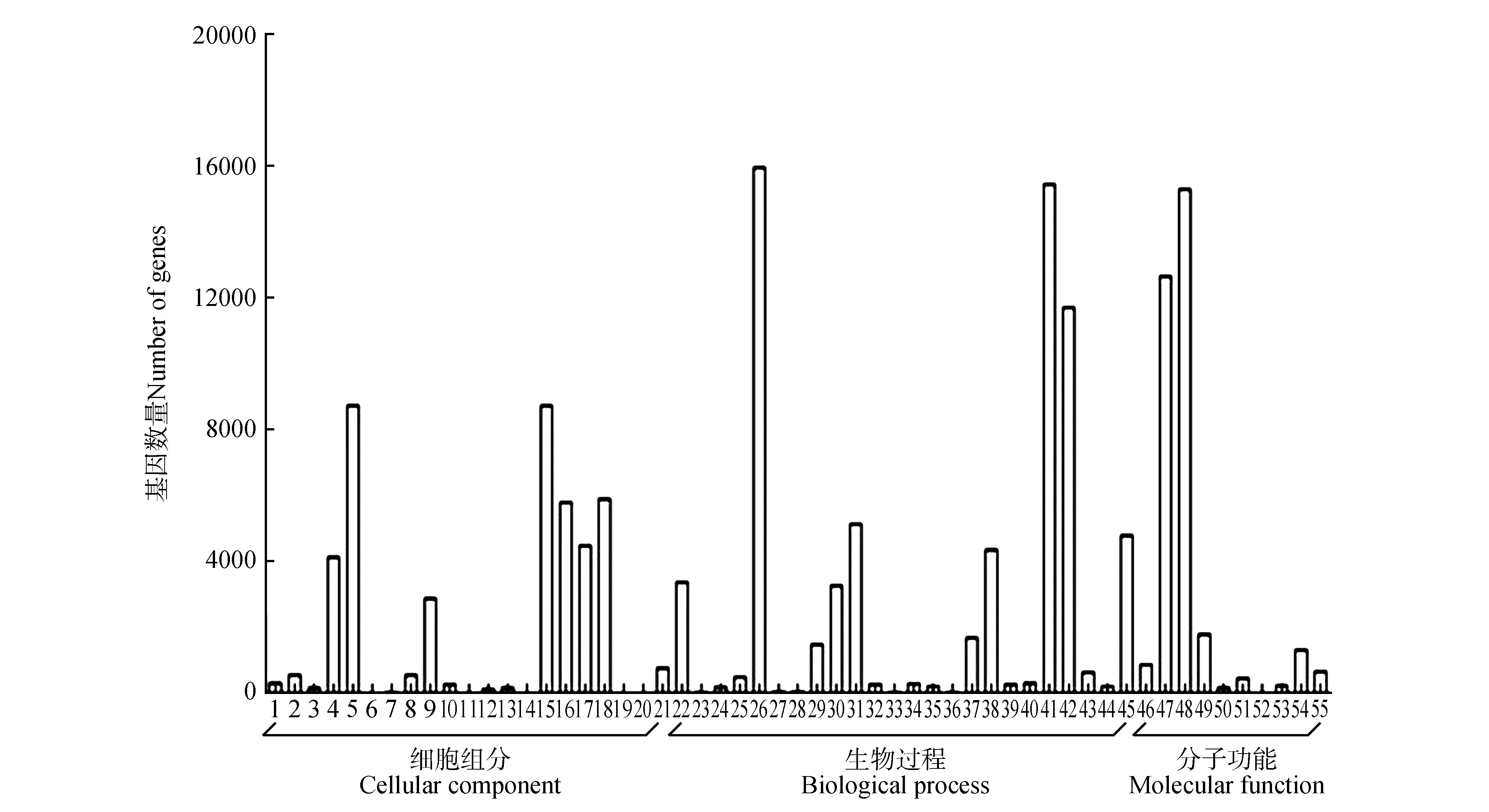

Fig.2
COG functional classification of unigenesA.RNA processing and modification;B.Chromatin structure and dynamics;C.Energy production and conversion;D.Cell cycle control, cell division, chromosome partitioning;E.Amino acid transport and metabolism;F.Nucleotide transport and metabolism;G.Carbohydrate transport and metabolism;H.Coenzyme transport and metabolism;I.Lipid transport and metabolism;J.Translation, ribosomal structure and biogenesis;K.Transcription;L.Replication, recombination and repair;M.Cell wall/membrane/envelope biogenesis;N.Cell motility;O.Posttranslational modification, protein turnover, chaperones;P.Inorganic ion transport and metabolism;Q.Secondary metabolites biosynthesis, transport and catabolism;R.General function prediction only;S.Function unknown;T.Signal transduction mechanisms;U.Intracellular trafficking, secretion, and vesicular transport;V.Defense mechanisms;W.Extracellular structures;Y.Nuclear structure;Z.Cytoskeleton
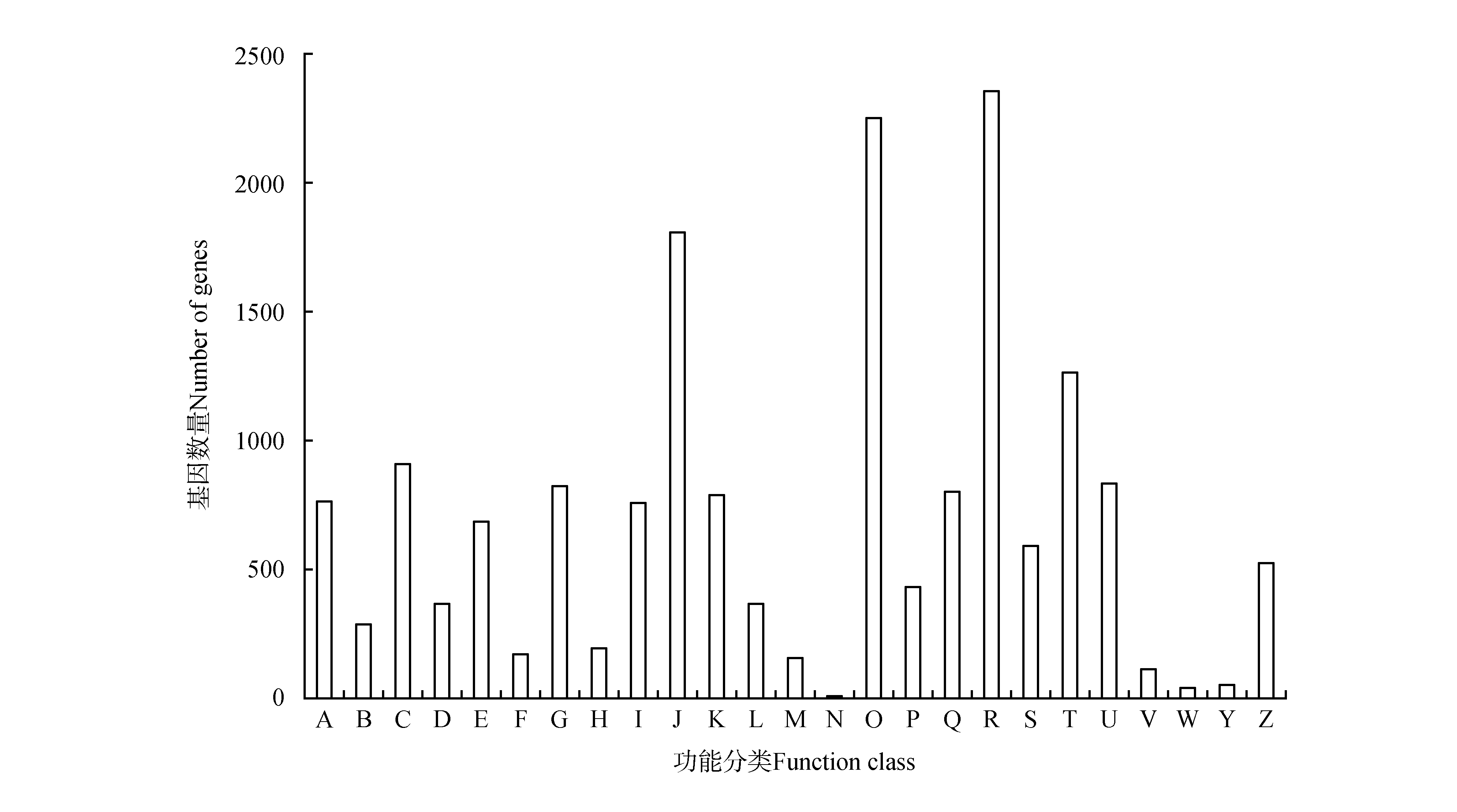
Table 3
Classification of unigenes KEGG metabolic pathways in P. tomentosa transcriptome
KEGG通路 KEGG pathway | 注释基因 Annotated genes | 占总基因数的比例 Percentage of total genes(%) | 通路ID Pathway ID |
|---|---|---|---|
| 核糖体Ribosome | 1 216 | 7.45 | ko03010 |
| 植物病原体相互作用Plant-bacterium warfare | 646 | 3.96 | ko04626 |
| 碳代谢Carbon metabolism | 638 | 3.91 | ko01200 |
| 氨基酸的生物合成Biosynthesis of amino acids | 559 | 3.43 | ko01230 |
| 内质网蛋白加工Protein processing in endoplasmic reticulum | 533 | 3.27 | ko04141 |
| 剪接Spliceosome | 396 | 2.43 | ko03040 |
| 淀粉和蔗糖代谢Starch and sucrose metabolism | 384 | 2.35 | ko00500 |
| RNA运输 RNA transport | 371 | 2.27 | ko03013 |
| 氧化磷酸化Oxidative phosphorylation | 370 | 2.27 | ko00190 |
| 植物激素信号转导Plant hormone signal transduction | 361 | 2.21 | ko04075 |
| 胞吞作用Endocytosis | 325 | 1.99 | ko04144 |
| 糖酵解/糖异生Glycolysis/gluconeogenesis | 298 | 1.83 | ko00010 |
| 嘌呤代谢Purine metabolism | 275 | 1.69 | ko00230 |
| 氨基糖和核苷酸糖代谢Amino sugar and nucleotide sugar metabolism | 268 | 1.64 | ko00520 |
| 苯丙素生物合成Phenylpropanoid biosynthesis | 266 | 1.63 | ko00940 |
| 泛素介导的蛋白水解Ubiquitin mediated proteolysis | 257 | 1.58 | ko04120 |
| 吞噬Phagocytosis | 248 | 1.52 | ko04145 |
| 丙酮酸代谢Pyruvate metabolism | 220 | 1.35 | ko00620 |
| RNA降解 RNA degradation | 220 | 1.35 | ko03018 |
| mRNA监视通路 mRNA surveillance pathway | 217 | 1.33 | ko03015 |
| 1 | Qiu Z B,Wan L C,Chen T,et al.The regulation of cambial activity in Chinese fir(Cunninghamia lanceolata)involves extensive transcriptome remodeling[J].New Phytologist,2013,199(3):708-719. |
| 2 | 许会敏,王莉,曹德昌,等.维管形成层活动周期调控研究进展[J].科学通报,2015,60(7):619-629. |
| Xu H M,Wang L,Cao D C,et al.Research progress on the regulation of cambium activity periodicity[J].Chinese Science Bulletin,2015,60(7):619-629. | |
| 3 | Hertzberg M,Aspeborg H,Schrader J,et al.A transcriptional roadmap to wood formation[J].Proceedings of the National Academy of Sciences of the United States of America,2001,98(25):14732-14737. |
| 4 | Gaete-Loyola J,Lagos C,Beltrán M F,et al.Transcriptome profiling of Eucalyptus nitens reveals deeper insight into the molecular mechanism of cold acclimation and deacclimation process[J].Tree Genetics & Genomes,2017,13(2):37. |
| 5 | 曹玉婷,莫家兴,欧阳磊,等.柳杉维管形成层及木质部区转录组序列及基因表达分析[J].分子植物育种,2019,17(3):762-770. |
| Cao Y T,Mo J X,Ouyang L,et al.Transcriptome sequence and gene expression in vascular cambium and xylem of Cryptomeria fortunei[J].Molecular Plant Breeding,2019,17(3):762-770. | |
| 6 | 杨光,梁坤南,黄桂华,等.基于高通量测序的柚木边材转录组分析[J].分子植物育种,2020,18(13):4274-4282. |
| Yang G,Liang K N,Huang G H,et al.Transcriptome analysis of sapwood xylem in the Teak(Tectona grandis L.F.) based on high-throughput sequencing[J].Molecular Plant Breeding,2020,18(13):4274-4282. | |
| 7 | Qiu Z B,He Y Y,Zhang Y M,et al.Genome-wide identification and profiling of microRNAs in Paulownia tomentosa cambial tissues in response to seasonal changes[J].Gene,2018,677:32-40. |
| 8 | Dong Y P,Fan G Q,Deng M J,et al.Genome-wide expression profiling of the transcriptomes of four Paulownia tomentosa accessions in response to drought[J].Genomics,2014,104(4):295-305. |
| 9 | 贾新平,孙晓波,邓衍明,等.鸟巢蕨转录组高通量测序及分析[J].园艺学报,2014,41(11):2329-2341. |
| Jia X P,Sun X B,Deng Y M,et al.Sequencing and analysis of the transcriptome of Asplenium nidus[J].Acta Horticulturae Sinica,2014,41(11):2329-2341. | |
| 10 | Grabherr M G,Haas B J,Yassour M,et al.Full-length transcriptome assembly from RNA-Seq data without a reference genome[J].Nature Biotechnology,2011,29(7):644-652. |
| 11 | Altschul S F,Gish W,Miller W,et al.Basic local alignment search tool[J].Journal of Molecular Biology,1990,215(3):403-410. |
| 12 | 许蔷薇,楼雄珍,杨彬,等.云锦杜鹃转录组分析[J].浙江农林大学学报,2019,36(6):1190-1198. |
| Xu Q W,Lou X Z,Yang B,et al.Transcriptome sequencing and analysis of Rhododendron fortunei[J].Journal of Zhejiang A&F University,2019,36(6):1190-1198. | |
| 13 | 马婧,邓楠,褚建民,等.泡泡刺高通量转录组鉴定及其黄酮类代谢途径初步分析[J].林业科学研究,2016,29(1):61-66. |
| Ma J,Deng N,Chu J M,et al.High-throughput transcriptome identification and flavonoids metabolic pathways in Nitraria sphaerocarpa[J].Forest Research,2016,29(1):61-66. | |
| 14 | 蔡年辉,邓丽丽,许玉兰,等.基于高通量测序的云南松转录组分析[J].植物研究,2016,36(1):75-83. |
| Cai N H,Deng L L,Xu Y L,et al.Transcriptome analysis for Pinus yunnanensis based on high throughput sequencing[J].Bulletin of Botanical Research,2016,36(1):75-83. | |
| 15 | 王帅,邵芬娟,李论,等.穗花杉的转录组测序及其转录组特性分析[J].林业科学研究,2017,30(5):759-764. |
| Wang S,Shao F J,Li L,et al.Transcriptome sequencing and analysis of Amentotaxus argotaenia(Hance) Pilger[J].Forest Research,2017,30(5):759-764. | |
| 16 | 李太强,刘雄芳,万友名,等.基于高通量测序的极小种群野生植物长梗杜鹃转录组分析[J].植物研究,2017,37(6):825-834. |
| Li T Q,Liu X F,Wan Y M,et al.Transcriptome analysis for Rhododendron longipedicellatum(plant species with extremely small populations) based on high throughput sequencing[J].Bulletin of Botanical Research,2017,37(6):825-834. | |
| 17 | 王兴春,谭河林,陈钊,等.基于RNA-Seq技术的连翘转录组组装与分析及SSR分子标记的开发[J].中国科学:生命科学,2015,45(3):301-310. |
| Wang X C,Tan H L,Chen Z,et al.Assembly and characterization of the transcriptome and development of SSR markers in Forsythia suspensa based on RNA-Seq technology[J].Scientia Sinica Vitae,2015,45(3):301-310. | |
| 18 | 林开勤,李岩,赵德刚.杜仲转录组SSR发掘及标记开发[J].分子植物育种,2016,14(6):1548-1558. |
| Lin K Q,Li Y,Zhao D G.SSR mining and marker development in Eucommia ulmoides oliver transcriptome[J].Molecular Plant Breeding,2016,14(6):1548-1558. | |
| 19 | 魏秀清,许玲,章希娟,等.莲雾转录组SSR信息分析及其分子标记开发[J].园艺学报,2018,45(3):541-551. |
| Wei X Q,Xu L,Zhang X J,et al.Analysis on SSR information in transcriptome and development of molecular markers in wax apple[J].Acta Horticulturae Sinica,2018,45(3):541-551. |
| [1] | Zhanmin ZHENG, Yubing SHANG, Guangbo ZHOU, Di XIAO, Yi LIU, Xiangling YOU. Genetic Transformation and Function Analysis of PsnHB13 and PsnHB15 of Populus simonii × Populus nigra [J]. Bulletin of Botanical Research, 2023, 43(3): 340-350. |
| [2] | Dongmei HUANG, Ying CHEN, Lu BAI, Di’an NI, Yiyang XU, Zhiguo ZHANG, Qiaoping QIN. Transcriptome Analysis of Hemerocallis fulva Leaves Respond to Low Temperature Stress [J]. Bulletin of Botanical Research, 2022, 42(3): 424-436. |
| [3] | Jingya Yu, Mingze Xia, Hao Xu, Faqi Zhang. Comparative Transcriptome Analysis of Three Artemisia Species in Qinghai Tibet Plateau [J]. Bulletin of Botanical Research, 2022, 42(2): 200-210. |
| [4] | Xue-Yan ZHAO, Si-Feng LI, Guo-Qing BAI, Wei-Min LI. Comparative Transcriptome Analysis of Five Species of Lilium [J]. Bulletin of Botanical Research, 2021, 41(4): 614-625. |
| [5] | Chen-Chen ZHOU, Yi-Liang LI, Zhe WANG, Fen-Cheng ZHAO, Sui-Ying ZHONG, Chang-Ming LIN, Zhi-Qiang TAN, Fu-Ming LI, Hui-Shan WU, Wen-Bing GUO, Fang-Yan LIAO, Jian-Ge WANG. Screening and Expression Analysis of Different Genes for Oleoresin Production in the Specific Period of Pinus elliottii×P.caribaea by RNA-Seq Technology [J]. Bulletin of Botanical Research, 2021, 41(3): 419-428. |
| [6] | Lin LU, Shang-Yu YANG, Wei-Dong LIU, Li-Ming LU. To Explore the Transcription Factor in Response to Low Temperature Stress in Nicotiana alata by Transcriptome Sequencing [J]. Bulletin of Botanical Research, 2021, 41(1): 119-129. |
| [7] | Li-Qiang ZHAO, Chun-Miao SHAN, Sheng-Xiang ZHANG, Yuan-Yuan SHI, Ke-Long MA, Jia-Wen WU. Identification of Key Enzyme Genes Involved in Anthocyanin Synthesis pathway in Clinopodium gracile by Transcriptome Analysis [J]. Bulletin of Botanical Research, 2020, 40(6): 886-896. |
| [8] | YANG Bin, MENG Qing-Yao, ZHANG Kai, DUAN Yi-Zhong. Chloroplast Genome Characterization and Identification of Genetic Relationship of Relict Endangered Plant Amygdalus nana [J]. Bulletin of Botanical Research, 2020, 40(5): 686-695. |
| [9] | LIU Dan, ZENG Qin-Meng, LIU Bin, LI Yu, CHEN Shi-Pin. Transcriptome Analysis and Gene Functional Annotation in Phoebe bournei [J]. Bulletin of Botanical Research, 2020, 40(4): 613-622. |
| [10] | ZHANG Yu, XIA Ming-Ze, ZHANG Fa-Qi. Transcriptome Analysis for Medicinal Plant Anisodus tanguticus [J]. Bulletin of Botanical Research, 2020, 40(3): 458-467. |
| [11] | ZHU Li-Li, DU Qing-Xin, HE Feng, QING Jun, DU Hong-Yan. Sequencing Analysis of Transcriptome of Male Floral Bud at Two Development Stages in Eucommia ulmoides [J]. Bulletin of Botanical Research, 2020, 40(2): 284-292. |
| [12] | DUAN Yi-Zhong, DU Zhong-Yu, WANG Hai-Tao. Chloroplast Genome Characteristics of Endangered Relict Plant Tetraena mongolica in the Arid Region of Northwest China [J]. Bulletin of Botanical Research, 2019, 39(5): 653-663. |
| [13] | YE Xing-Zhuang, LIU Dan, LUO Jia-Jia, FAN Hui-Hua, ZHANG Guo-Fang, LIU Bao, CHEN Shi-Pin. Transcriptome Analysis for Rare and Endangered Plants of Semiliquidambar cathayensis [J]. Bulletin of Botanical Research, 2019, 39(2): 276-286. |
| [14] | LIU Xiong-Fang, LI Tai-Qiang, ZHANG Xu, LI Zheng-Hong, WAN You-Ming, AN Jing, LIU Xiu-Xian, MA Hong. Characteristic Analysis of Microsatellites in the Transcriptome of Phyllanthus emblica,an Important Edible and Medicinal Plant [J]. Bulletin of Botanical Research, 2019, 39(2): 294-302. |
| [15] | WANG Da-Wei, ZHOU Fan, SHEN Bing-Qi, WANG Lian-Chun. Characteristics of Microsatellite in Camellia saluenensis by High-throughput Sequencing [J]. Bulletin of Botanical Research, 2019, 39(1): 148-155. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||