Bulletin of Botanical Research ›› 2021, Vol. 41 ›› Issue (1): 107-118.doi: 10.7525/j.issn.1673-5102.2021.01.014
Previous Articles Next Articles
Meng-Xuan REN1, Yang ZHANG1, Shuang WANG1, Rui-Qi WANG1, Cong LIU1, Zhi-Gang WEI2( )
)
Received:2020-06-02
Online:2021-01-20
Published:2021-01-05
Contact:
Zhi-Gang WEI
E-mail:zhigangwei1973@163.com
About author:REN Meng-Xuan(1992—),male,master,mainly engaged in plant secondary cell wall research.
Supported by:CLC Number:
Meng-Xuan REN, Yang ZHANG, Shuang WANG, Rui-Qi WANG, Cong LIU, Zhi-Gang WEI. Genome-wide Identification and Expression Analysis GATA Family of Populus trichocarpa[J]. Bulletin of Botanical Research, 2021, 41(1): 107-118.
Add to citation manager EndNote|Ris|BibTeX
URL: https://bbr.nefu.edu.cn/EN/10.7525/j.issn.1673-5102.2021.01.014
Table 1
Primers for quantitative real-time PCR of P.trichocarpa GATA family
基因名字 Gene name | 登录号 Accession number | 引物名称 Primer name | 引物序列 Primer sequence(5′-3′) |
|---|---|---|---|
| PtrGATA1.1 | Potri.001G053500.1 | DL-Ptr001G053500-F DL-Ptr001G053500-R | AGACACGGCTGCTTGTTTCA GCCCTTCTGGATTTTTTGCT |
| PtrGATA1.2 | Potri.001G151700.1 | DL-Ptr001G151700-F DL-Ptr001G151700-R | TTGCTCTGGATTACAACGAAGG TCCGAGTTGGATATGGCTGAC |
| PtrGATA1.3 | Potri.001G188500.1 | DL-Ptr001G188500-F DL-Ptr001G188500-R | CCAAACCAACATCCACATCCT CATTTTAGTGGTGGATGGGACA |
| PtrGATA2.1 | Potri.002G110800.1 | DL-Ptr002G110800-F DL-Ptr002G110800-R | CGTTTTCCCTGCTGTTACCC AACTGCCCATTCTTGCGG |
| PtrGATA2.2 | Potri.002G110900.1 | DL-Ptr002G110900-F DL-Ptr002G110900-R | AACCTCAACGAGCAGCTTCA TGACGGACACCATATCTAACTTTC |
| PtrGATA2.3 | Potri.002G142800.1 | DL-Ptr002G142800-F DL-Ptr002G142800-R | AGTGATGACGTGGCAGAGCTA TGCTACAAGATTTGATGCGAGA |
| PtrGATA2.4 | Potri.002G199800.1 | DL-Ptr002G199800-F DL-Ptr002G199800-R | ATCAGACAGAGGAAGAGGAGGAG CCACAAGATAAAGCCATCAAGAG |
| PtrGATA3.1 | Potri.003G082800.1 | DL-Ptr003G082800-F DL-Ptr003G082800-R | TATGACAATAGGGTTGCTCTGGA ACAACTGATTGGGCTGGACC |
| PtrGATA3.2 | Potri.003G174800.1 | DL-Ptr003G174800-F DL-Ptr003G174800-R | GCCCAGTCTCCGTTCTTGA TCTCCCCATTTTTGCTACTGAC |
| PtrGATA3.3 | Potri.003G213300.1 | DL-Ptr003G213300-F DL-Ptr003G213300-R | TGCTTACCCCAAGTCCTACAGT GCTACTGTGTTGAGGAGTGTTCG |
| PtrGATA4.1 | Potri.004G161500.1 | DL-Ptr004G161500-F DL-Ptr004G161500-R | GCTTGAATGGGTATCTCACTTTG AACCAGGAGTTTTTGCGAGAG |
| PtrGATA4.2 | Potri.004G211800.1 | DL-Ptr004G211800-F DL-Ptr004G211800-R | CAAGCAGAAGCAAACCCATC TTGTCGCATTTTCGGTGG |
| PtrGATA5.1 | Potri.005G020500.1 | DL-Ptr005G020500-F DL-Ptr005G020500-R | ATCACTACAGCCACAACAACAAAC TCCTTTGGCACCTTTGTTTCT |
| PtrGATA5.2 | Potri.005G066100.1 | DL-Ptr005G066100-F DL-Ptr005G066100-R | TGGATAGGCAGTGAACTTAGCG TCCCAAAAAATCCCCAAAAT |
| PtrGATA5.3 | Potri.005G117600.1 | DL-Ptr005G117600-F DL-Ptr005G117600-R | TGAACTTTGTGTGCCGACG ACAAGGCTCTGTTAGGACCGA |
| PtrGATA5.4 | Potri.005G122700.1 | DL-Ptr005G122700-F DL-Ptr005G122700-R | ATCCCCCTCAAGCCCATAA TTGCCCTTCTCTCTTCCTTCTT |
| PtrGATA5.5 | Potri.005G152500.1 | DL-Ptr005G152500-F DL-Ptr005G152500-R | TTTCCCTGCTGTTACCCCA CACGGAATCTAACAAGTGAGGC |
| PtrGATA5.6 | Potri.005G152800.1 | DL-Ptr005G152800-F DL-Ptr005G152800-R | TGGAAATGACGCCTCAAAAC AACCTCCTGACGGACACCATA |
| PtrGATA6.1 | Potri.006G229200.1 | DL-Ptr006G229200-F DL-Ptr006G229200-R | AACCAAACCTGGAGAGTCGC GAACTTTCACCACTTTCTTCAGCT |
| PtrGATA6.2 | Potri.006G237700.1 | DL-Ptr006G237700-F DL-Ptr006G237700-R | CCACTGTGACTTTCGTTGATAGC TCATCATACGGCACGCAAA |
| PtrGATA7.1 | Potri.007G016600.1 | DL-Ptr007G016600-F DL-Ptr007G016600-R | AGGAGGAGGGGGGAGATAAA CGTCGGCACACAAAGTTCAC |
| PtrGATA7.2 | Potri.007G024500.1 | DL-Ptr007G024500-F DL-Ptr007G024500-R | CCACCTCAAGCCCATAAAACT TCTCTCTTCCTTTTTGAACCGA |
| PtrGATA7.3 | Potri.007G116600.1 | DL-Ptr007G116600-F DL-Ptr007G116600-R | TGGTCCAAGTGGTCCAAGAA GCCGCAGTTATGGTATCAGAAT |
| PtrGATA7.4 | Potri.007G116700.1 | DL-Ptr007G116700-F DL-Ptr007G116700-R | TGGGAATGATGGCGTGTTT GCATTCCTGGCATTCTGTTG |
| PtrGATA8.1 | Potri.008G038900.1 | DL-Ptr008G038900-F DL-Ptr008G038900-R | GAAGCCTGACGATGAAGAAAGA CTCAAGGACAGAAACTGGGCT |
| PtrGATA8.2 | Potri.008G213900.1 | DL-Ptr008G213900-F DL-Ptr008G213900-R | AAAGGGGCAGCAAATGACA AGACCCATAAGACAACGCCAT |
| PtrGATA9.1 | Potri.009G123400.1 | DL-Ptr009G123400-F DL-Ptr009G123400-R | TTCGTGGATGATTCCGTGTC CAAGAAGTTTTTGCGAAAGTAGG |
| PtrGATA10.1 | Potri.010G001300.1 | DL-Ptr010G001300-F DL-Ptr010G001300-R | ATTAGGGTTGAATAAGGGAGGG TTGAACCAAGACTTCTCTACCCA |
| PtrGATA10.2 | Potri.010G223300.1 | DL-Ptr010G223300-F DL-Ptr010G223300-R | CATGGACGAGATAGATTGTGGAA TGAGACGACCAAATACCAGGAA |
| PtrGATA10.3 | Potri.010G251600.1 | DL-Ptr010G251600-F DL-Ptr010G251600-R | TTTTTGACTCTGTTTCACCTGGA CCTGACAGCCCCCTATTATTACT |
| PtrGATA13.1 | Potri.013G059600.1 | DL-Ptr013G059600-F DL-Ptr013G059600-R | TACAGAAGAAAACCCCCCAAA CTGAAGCAAAGGAGGGTCTGA |
| PtrGATA14.1 | Potri.014G058600.1 | DL-Ptr014G058600-F DL-Ptr014G058600-R | CCACTGACTTCACTGACCATCTC TGAGCGTTTGCTGCGAGA |
| PtrGATA14.2 | Potri.014G124400.1 | DL-Ptr014G124400-F DL-Ptr014G124400-R | GAGCGGAGAGGAGTAAAAATAAGA CGGACCAAGCCACAAGATAA |
| PtrGATA17.1 | Potri.017G042200.1 | DL-Ptr017G042200-F DL-Ptr017G042200-R | GAGAGCCTGAGACATCCACAAT ACCTTTCCAGGGAGGCAAT |
| PtrGATA17.2 | Potri.017G042300.1 | DL-Ptr017G042300-F DL-Ptr017G042300-R | AATCGGTTCAAAGTCCACTCC CCTCTACCTGCAGTAAGTTCACAA |
| PtrGATA18.1 | Potri.018G044900.1 | DL-Ptr018G044900-F DL-Ptr018G044900-R | GCTTTGCGTGCCCTATGAT TCTTGGATGATTCGTCTGGC |
| PtrGATA18.2 | Potri.018G053600.1 | DL-Ptr018G053600-F DL-Ptr018G053600-R | TCATCGAAAACGGAGGACG GTTTCTGAGCGATTTGAGTTGG |
| PtrGATA19.1 | Potri.019G033000.1 | DL-Ptr019G033000-F DL-Ptr019G033000-R | AAAACCCCCCAAAACCCT GGAGCAAAGGAGGGTCTGAG |
| PtrGATAT.1 | Potri.T158300.1 | DL-PtrT158300-F DL-PtrT158300-R | CTCTCATCATGACGTTCACTTAGG TCTTGGATGATTCGTCTGGC |
Table 2
Summary of GATA genes in P.trichocarpa
基因号 Gene ID | 基因名字 Gene name | 基因的位置 Genomic position | 长度 Length(aa) | 分子量 MW(Da) | 等电点 pl | CDs长度 Length of CDs(bp) |
|---|---|---|---|---|---|---|
| Potri.001G053500.1 | PtrGATA1.1 | Chr01:4086401..4088825 reverse | 336 | 38 285.85 | 9.68 | 1 011 |
| Potri.001G151700.1 | PtrGATA1.2 | Chr01:12502479..12509399 reverse | 552 | 61 305.85 | 6.96 | 1 659 |
| Potri.001G188500.1 | PtrGATA1.3 | Chr01:16658546..16659881 reverse | 250 | 28 010.26 | 5.41 | 753 |
| Potri.002G110800.1 | PtrGATA2.1 | Chr02:8198186..8203728 reverse | 360 | 39 278.43 | 4.87 | 1 083 |
| Potri.002G110900.1 | PtrGATA2.2 | Chr02:8206492..8211812 reverse | 290 | 31 465.78 | 5.93 | 873 |
| Potri.002G142800.1 | PtrGATA2.3 | Chr02:10597460..10598816 forward | 246 | 27 390.13 | 6.03 | 741 |
| Potri.002G199800.1 | PtrGATA2.4 | Chr02:16060278..16061389 forward | 147 | 16 627.16 | 9.40 | 444 |
| Potri.003G082800.1 | PtrGATA3.1 | Chr03:11053999..11060297 forward | 540 | 60 284.54 | 6.58 | 1 623 |
| Potri.003G174800.1 | PtrGATA3.2 | Chr03:18351164..18353616 forward | 259 | 29 021.80 | 9.17 | 780 |
| Potri.003G213300.1 | PtrGATA3.3 | Chr03:21117706..21119661 reverse | 226 | 25 422.98 | 8.74 | 681 |
| Potri.004G161500.1 | PtrGATA4.1 | Chr04:18221923..18223879 forward | 327 | 36 306.48 | 8.53 | 984 |
| Potri.004G211800.1 | PtrGATA4.2 | Chr04:21951641..21956060 reverse | 301 | 33 973.34 | 8.57 | 906 |
| Potri.005G020500.1 | PtrGATA5.1 | Chr05:1593397..1595249 forward | 161 | 17 587.24 | 9.78 | 486 |
| Potri.005G066100.1 | PtrGATA5.2 | Chr05:4766766..4769587 reverse | 356 | 29 329.51 | 8.75 | 771 |
| Potri.005G117600.1 | PtrGATA5.3 | Chr05:9096540..9098179 reverse | 333 | 36 840.45 | 7.10 | 1 002 |
| Potri.005G122700.1 | PtrGATA5.4 | Chr05:9549440..9551215 reverse | 254 | 28 599.41 | 7.55 | 765 |
| Potri.005G152500.1 | PtrGATA5.5 | Chr05:14348124..14353280 reverse | 365 | 39 708.23 | 5.13 | 1 098 |
| Potri.005G152800.1 | PtrGATA5.6 | Chr05:14386485..14398559 reverse | 288 | 31 463.70 | 5.67 | 867 |
| Potri.006G229200.1 | PtrGATA6.1 | Chr06:24012190..24015009 forward | 355 | 39 445.47 | 9.04 | 1 068 |
| Potri.006G237700.1 | PtrGATA6.2 | Chr06:24697287..24699200 forward | 373 | 41 397.57 | 6.04 | 1 122 |
| Potri.007G016600.1 | PtrGATA7.1 | Chr07:1263957..1265632 reverse | 376 | 41 808.29 | 6.95 | 1 131 |
| Potri.007G024500.1 | PtrGATA7.2 | Chr07:1849537..1850974 reverse | 254 | 28 668.60 | 8.46 | 765 |
| Potri.007G116600.1 | PtrGATA7.3 | Chr07:13732111..13734218 forward | 142 | 14 882.95 | 8.47 | 429 |
| Potri.007G116700.1 | PtrGATA7.4 | Chr07:13736385..13742259 forward | 384 | 43 220.87 | 4.90 | 1 155 |
| Potri.008G038900.1 | PtrGATA8.1 | Chr08:2194430..2198138 forward | 354 | 38 941.87 | 6.52 | 1 065 |
| Potri.008G213900.1 | PtrGATA8.2 | Chr08:16937468..16938757 reverse | 138 | 14 783.70 | 9.69 | 417 |
| Potri.009G123400.1 | PtrGATA9.1 | Chr09:10283234..10285131 forward | 329 | 36 593.74 | 6.61 | 990 |
| Potri.010G001300.1 | PtrGATA10.1 | Chr10:149903..150769 reverse | 153 | 16 419.65 | 9.85 | 462 |
| Potri.010G223300.1 | PtrGATA10.2 | Chr10:20747463..20750870 reverse | 446 | 49 067.91 | 5.93 | 1 341 |
| Potri.010G251600.1 | PtrGATA10.3 | Chr10:22342132..22345674 forward | 307 | 33 369.72 | 7.80 | 924 |
| Potri.013G059600.1 | PtrGATA13.1 | Chr13:4460579..4462220 forward | 295 | 32 672.55 | 6.53 | 888 |
| Potri.014G058600.1 | PtrGATA14.1 | Chr14:4540248..4541744 forward | 251 | 27 963.80 | 6.42 | 756 |
| Potri.014G124400.1 | PtrGATA14.2 | Chr14:9595039..9596182 forward | 133 | 14 704.83 | 9.77 | 402 |
| Potri.017G042200.1 | PtrGATA17.1 | Chr17:3570422..3576737 reverse | 406 | 46 027.48 | 5.36 | 1 221 |
| Potri.017G042300.1 | PtrGATA17.2 | Chr17:3583593..3583948 reverse | 115 | 12 867.23 | 9.94 | 348 |
| Potri.018G044900.1 | PtrGATA18.1 | Chr18:4078772..4080396 reverse | 380 | 42 109.26 | 6.22 | 1 143 |
| Potri.018G053600.1 | PtrGATA18.2 | Chr18:5587016..5588879 forward | 303 | 33 666.66 | 8.78 | 912 |
| Potri.018G033000.1 | PtrGATA19.1 | Chr19:3788859..3790666 reverse | 294 | 32 198.69 | 6.18 | 885 |
| Potri.T158300.1 | PtrGATAT.1 | scaffold_694:4641..6156 forward | 354 | 39 505.39 | 6.19 | 1 065 |
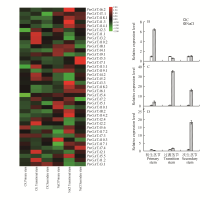
Fig.6
The expression pattern analysis of GATA genes in stem development in P.trichocarpa under salt stressA.Color temperature map of the expression level of GATA gene in the stem of P.trichocarpa under salt stress;B.PtrGATA10.1 stem expression under salt stress;C.PtrGATA18.1 stem expression level under salt stress;D.PtrGATA5.3 Stem expression level under salt stress

| 1 | 靳进朴,郭安源,何坤,等.植物转录因子分类、预测和数据库构建[J].生物技术通报,2015,31(11):68-77. |
| Jin J P,Guo A Y,He K,et al.Classification,prediction and database construction of plant transcription factors[J].Biotechnology Bulletin,2015,31(11):68-77. | |
| 2 | Franco-Zorrilla J M,Solano R.High-throughput analysis of protein-DNA binding affinity[M].//Sanchez-Serrano J,Salinas J.Arabidopsis Protocols.Methods in Molecular Biology(Methods and Protocols).Totowa,NJ:Humana Press,2014,1062:697-709. |
| 3 | Riechmann J L,Heard J,Martin G,et al.Arabidopsis transcription factors:genome-wide comparative analysis among eukaryotes[J].Science,2000,290(5499):2105-2110. |
| 4 | Evans T,Reitman M,Felsenfeld G.An erythrocyte-specific DNA-binding factor recognizes a regulatory sequence common to all chicken globin genes[J].Proceedings of the National Academy of Sciences of the United States of America,1988,85(16):5976-5980. |
| 5 | 史莹华,许梓荣.GATA转录因子研究进展[J].生物学通报,2005,40(3):1-2. |
| Shi Y H,Xu Z R.Recent advances in GATA transcription factor[J].Bulletin of Biology,2005,40(3):1-2. | |
| 6 | Daniel-Vedele F,Caboche M.A tobacco cDNA clone encoding a GATA-1 zinc finger protein homologous to regulators of nitrogen metabolism in fungi[J].Molecular and General Genetics MGG,1993,240(3):365-373. |
| 7 | Reyes J C,Muro-Pastor M I,Florencio F J.The GATA family of transcription factors in Arabidopsis and rice[J].Plant Physiology,2004,134(4):1718-1732. |
| 8 | 敖涛,廖晓佳,徐伟,等.蓖麻GATA基因家族的鉴定和特征分析[J].植物分类与资源学报,2015,37(4):453-462. |
| Ao T,Liao X J,Xu W,et al.Identification and characterization of GATA gene family in castor bean(Ricinus communis)[J].Plant Diversity and Resources,2015,37(4):453-462. | |
| 9 | 陈鸿飞.苹果CCO和GATA基因家族的鉴定、系统进化和非生物胁迫的表达分析[D].杨凌:西北农林科技大学,2018. |
| Chen H F.Genome-wide identification,phylogenetic evolution and expression analysis of CCO and GATA gene families in apple[D].Yangling:Northwest A & F University,2018. | |
| 10 | 骆鹰,王有成,王伟平,等.水稻GATA基因家族生物信息学分析[J].分子植物育种,2018,16(17):5514-5522. |
| Luo Y,Wang Y C,Wang W P,et al.Bioinformatics analysis of GATA gene family in rice[J].Molecular Plant Breeding,2018,16(17):5514-5522. | |
| 11 | 袁岐.番茄GATA转录因子的全基因组挖掘及抗逆相关基因筛选[D].哈尔滨:东北农业大学,2018. |
| Yuan Q.Whole-genome mining of GATA transcription factor from tomato and screening of resistance-related genes[D].Harbin:Northeast Agricultural University,2018. | |
| 12 | 袁岐,张春利,赵婷婷,等.辣椒GATA转录因子的生物信息学分析[J].中国农学通报,2017,33(17):24-31. |
| Yuan Q,Zhang C L,Zhao T T,et al.Bioinformatics analysis of GATA transcription factor in pepper[J].Chinese Agricultural Science Bulletin,2017,33(17):24-31. | |
| 13 | Shokrollahi B,Baneh H.(Co)variance components and genetic parameters for growth traits in Arabi sheep using different animal models[J].Genetics and Molecular Research,2012,11(1):305-314. |
| 14 | Hudson D,Guevara D R,Hand A J,et al.Rice cytokinin GATA transcription factor1 regulates chloroplast development and plant architecture[J].Plant Physiology,2013,162(1):132-144. |
| 15 | Hudson D,Guevara D,Yaish M W,et al.GNC and CGA1 modulate chlorophyll biosynthesis and glutamate synthase(GLU1/Fd-GOGAT) expression in Arabidopsis[J].PLoS One,2011,6(11):e26765. |
| 16 | An Y,Han X,Tang S,et al.Poplar GATA transcription factor PdGNC is capable of regulating chloroplast ultrastructure,photosynthesis,and vegetative growth in Arabidopsis under varying nitrogen levels[J].Plant Cell,Tissue and Organ Culture,2014,119(2):313-327. |
| 17 | Kiba T,Naitou T,Koizumi N,et al.Combinatorial microarray analysis revealing Arabidopsis genes implicated in cytokinin responses through the His→Asp phosphorelay circuitry[J].Plant and Cell Physiology,2005,46(2):339-355. |
| 18 | Naito T,Kiba T,Koizumi N,et al.Characterization of a unique GATA family gene that responds to both light and cytokinin in Arabidopsis thaliana[J].Bioscience,Biotechnology,and Biochemistry,2007,71(6):1557-1560. |
| 19 | Mara C D,Irish V F.Two GATA transcription factors are downstream effectors of floral homeotic gene action in Arabidopsis[J].Plant Physiology,2008,147(2):707-718. |
| 20 | Liu P P,Koizuka N,Martin R C,et al.The BME3(Blue Micropylar End 3) GATA zinc finger transcription factor is a positive regulator of Arabidopsis seed germination[J].The Plant Journal,2005,44(6):960-971. |
| 21 | Jansson S,Douglas C J.Populus:a model system for plant biology[J].Annual Review of Plant Biology,2007,58:435-458. |
| 22 | Tuskan G A,Difazio S,Jansson S,et al.The genome of black cottonwood,Populus trichocarpa(Torr.&Gray)[J].Science,2006,313(5793):1596-1604. |
| 23 | 王爽,张洋,任梦轩,等.全基因组分析杨树WOX基因家族在茎部发育中的作用[J].植物研究,2019,39(4):568-577. |
| Wang S,Zhang Y,Ren M X,et al.Genome-wide analysis of the WOX family reveals their involvement in stem growth of Populus trichocarpa[J].Bulletin of Botanical Research,2019,39(4):568-577. | |
| 24 | Mukhopadhyay A,Vij S,Tyagi A K.Overexpression of a zinc-finger protein gene from rice confers tolerance to cold,dehydration,and salt stress in transgenic tobacco[J].Proceedings of the National Academy of Sciences of the National Academy of Sciences,2004,101(16):6309-6314. |
| 25 | Huang X Y,Chao D Y,Gao J P,et al.A previously unknown zinc finger protein,DST,regulates drought and salt tolerance in rice via stomatal aperture control[J].Genes & Development,2009,23(15):1805-1817. |
| 26 | Davletova S,Schlauch K,Coutu J,et al.The zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis[J].Plant Physiology,2005,139(2):847-856. |
| [1] | Lei XU, Xiao XU, Qinsong LIU. Effects of Exogenous Salicylic Acid on Antioxidant System and Gene Expression of Davidia involucrata Seedlings under Salt Stress [J]. Bulletin of Botanical Research, 2023, 43(4): 572-581. |
| [2] | Haonan ZHANG, Shanshan CHEN, Jianmin XU, Ping LUO, Xiaoping WANG, Zhiru XU, Chunjie FAN. Cloning and Functional Analysis of EgrWAT1 Gene in Eucalyptus grandis [J]. Bulletin of Botanical Research, 2023, 43(4): 601-611. |
| [3] | Li LI, Xin WANG, Yuejing ZHANG, Lingyun JIA, Hailong PANG, Hanqing FENG. Effects of Abiotic Stresses on the Intracellular and Extracellular ATP Levels of Tobacco Suspension Cells [J]. Bulletin of Botanical Research, 2023, 43(2): 179-185. |
| [4] | Shixian LIAO, Yuting WANG, Liben DONG, Yongmei GU, Fenglin JIA, Tingbo JIANG, Boru ZHOU. Function Analysis of the Transcription Factor PsnbZIP1 of Populus simonii×P. nigra in Response to Salt Stress [J]. Bulletin of Botanical Research, 2023, 43(2): 288-299. |
| [5] | Senyao LIU, Fenglin JIA, Qing GUO, Gaofeng FAN, Boru ZHOU, Tingbo JIANG. Response Analysis of Transcription Factor PsnbHLH162 Gene in Populus simonii × P. nigra under Salt Stress and Low Temperature Stress [J]. Bulletin of Botanical Research, 2023, 43(2): 300-310. |
| [6] | Anying HUANG, Dean XIA, Yang ZHANG, Dongchen NA, Qing YAN, Zhigang WEI. Cloning and Drought Tolerance Expression Analysis of PtrWRKY51 Gene in Populus trichocarpa [J]. Bulletin of Botanical Research, 2022, 42(6): 1005-1013. |
| [7] | Denggao LI, Rui LIN, Qinghui MU, Na ZHOU, Yanru ZHANG, Wei BAI. Identification and Analysis of the Potato StCRKs Gene Family and Expression Patterns in Response to Stress Signals [J]. Bulletin of Botanical Research, 2022, 42(6): 1033-1043. |
| [8] | Liran YUE, Yingjie LIU, Chenxu LIU, Yunwei ZHOU. Cloning and Functional Analysis of miR398a from Chrysanthemum× grandiflora in Response to Salt Stress [J]. Bulletin of Botanical Research, 2022, 42(6): 986-996. |
| [9] | Denggao LI, Rui LIN, Qinghui MU, Na ZHOU, Yanru ZHANG, Wei BAI. Cloning and Functional Analysis of StNPR4 gene in Solanum tuberosum [J]. Bulletin of Botanical Research, 2022, 42(5): 821-829. |
| [10] | Hongpeng WANG, Yidan LI, Yao WANG, Xiaoyu TAN, Chengbin CHEN, Lipeng ZHANG. Gene Cloning and Interaction Proteins Screening of DcPMK in Dioscorea composite [J]. Bulletin of Botanical Research, 2022, 42(5): 855-865. |
| [11] | Xueying WANG, Ruiqi WANG, Yang ZHANG, Cong LIU, Dean XIA, Zhigang WEI. Genome‑wide Identification and Stress Response Analysis of Cyclic Nucleotide-gated Channels(CNGC) Gene Family in Populus trichocarpa [J]. Bulletin of Botanical Research, 2022, 42(4): 613-625. |
| [12] | Jiaorao CHEN, Xu XU, Zhangli HU, Shuang YANG. Recent Advances on Salt Stress Sensitivity and Related Calcium Signals in Plants [J]. Bulletin of Botanical Research, 2022, 42(4): 713-720. |
| [13] | He CHENG, Shuanghui TIAN, Yang ZHANG, Cong LIU, De’an XIA, Zhigang WEI. Genome-wide Identification and Expression Analysis of nsLTP Gene Family in Populus trichocarpa [J]. Bulletin of Botanical Research, 2022, 42(3): 412-423. |
| [14] | Qian Sun, Yuhang Wu, Yaxuan Zhang, Jingdan Cao, Jingjing Shi, Chao Wang. Bioinformatic Analysis and Expression Pattern of LTP Family Genes in Populus davidiana × P. alba var. pyramidalis [J]. Bulletin of Botanical Research, 2022, 42(2): 211-223. |
| [15] | Shuang Ma, Boya Wang, Ying Cao, Shanglian Hu, Zhimin Gao. Identification and Expression Analysis of Expansin Genes in Moso Bamboo (Phyllostachys edulis) [J]. Bulletin of Botanical Research, 2022, 42(1): 29-38. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||