Bulletin of Botanical Research ›› 2021, Vol. 41 ›› Issue (4): 614-625.doi: 10.7525/j.issn.1673-5102.2021.04.017
• Research report • Previous Articles Next Articles
Xue-Yan ZHAO, Si-Feng LI, Guo-Qing BAI, Wei-Min LI( )
)
Received:2020-08-29
Online:2021-07-20
Published:2021-03-24
Contact:
Wei-Min LI
E-mail:763730163@qq.com
About author:ZHAO Xue-Yan(1986—),female,lecturer,doctor,Research interests: anatomical structure of plants and resource exploitation and utilization.
Supported by:CLC Number:
Xue-Yan ZHAO, Si-Feng LI, Guo-Qing BAI, Wei-Min LI. Comparative Transcriptome Analysis of Five Species of Lilium[J]. Bulletin of Botanical Research, 2021, 41(4): 614-625.
Add to citation manager EndNote|Ris|BibTeX
URL: https://bbr.nefu.edu.cn/EN/10.7525/j.issn.1673-5102.2021.04.017
Table 1
Statistical information for transcriptome seque-ncing of four species of Lilium
细叶百合 L. pumilum | 川百合 L. davidii | 卷丹 L. lancifolium | 百合 L. brownii var. viridulum | |
|---|---|---|---|---|
组装的总碱基 Total assembled bases | 31 251 564 | 37 017 020 | 28 657 033 | 32 992 838 |
Unigene数目 Unigene number | 44 565 | 51 413 | 41 638 | 44 716 |
GC含量 GC percentage(%) | 48.25 | 47.91 | 47.34 | 47.76 |
| N50 | 1 066 | 1 149 | 1 017 | 1 183 |
平均长度 Average length | 701 | 719 | 688 | 737 |
Table 2
Annotation information of five species of Lilium
野百合 L. brownii | 细叶百合 L. pumilum | 川百合 L. davidii | 卷丹 L. lancifolium | 百合 L. brownii var. viridulum | |
|---|---|---|---|---|---|
| Nr | 28 104 | 28 681 | 31 641 | 26 438 | 29 932 |
| Swiss-prot | 17 739 | 18 283 | 20 390 | 17 381 | 19 951 |
| GO | 6 494 | 6 785 | 7 398 | 6 475 | 7 360 |
| KOG | 14 682 | 14 958 | 16 619 | 14 371 | 16 268 |
| KEGG | 11 984 | 12 557 | 13 570 | 11 960 | 13 247 |
所有注释的基因数目 All annotation unigenes | 28 187 | 28 709 | 31 827 | 26 527 | 30 025 |
注释基因的百分比 The percentage of annotated unigenes(%) | 68.89 | 55.23 | 61.54 | 56.96 | 48.93 |
Fig.1
Comparison of gene ontogy(GO) functional classification of five species of Lilium1.Metabolic process;2.Cellular process;3.Single-organism process;4.Biological regulation;5.Localization;6.Regulation of biological process;7.Response to stimulus;8.Cellular component organization or biogenesis;9.Developmental process;10.Multicellular organismal process;11.Signaling;12.Reproduction;13.Reproductive process;14.Multi-organism process;15.Negative regulation of biological process;16.Positive regulation of biological process;17.Growth;18.Immune system process;19.Biological adhesion;20.Rhythmic process;21.Detoxification;22.Cell killing;23.Locomotion;24.Cell;25.Cell part;26.Organelle;27.Membrane;28.Membrane part;29.Organelle part;30.Macromolecular complex;31.Cell junction;32.Extracellular region;33.Membrane-enclosed lumen;34.Virion;35.Virion part;36.Extracellular matrix;37.Nucleoid;38.Extracellular matrix component part;39.Extracellular region part;40.Supramolecular fiber;41.Catalytic activity;42.Binding;43.Transporter activity;44.Structural molecule activity;45.Nucleic acid binding transcription factor activity;46.Molecular transducer activity;47.Signal transducer activity;48.Molecular function regulator;49.Antioxidant activity;50.Transcription factor activity, protein binding;51.Electron carrier activity
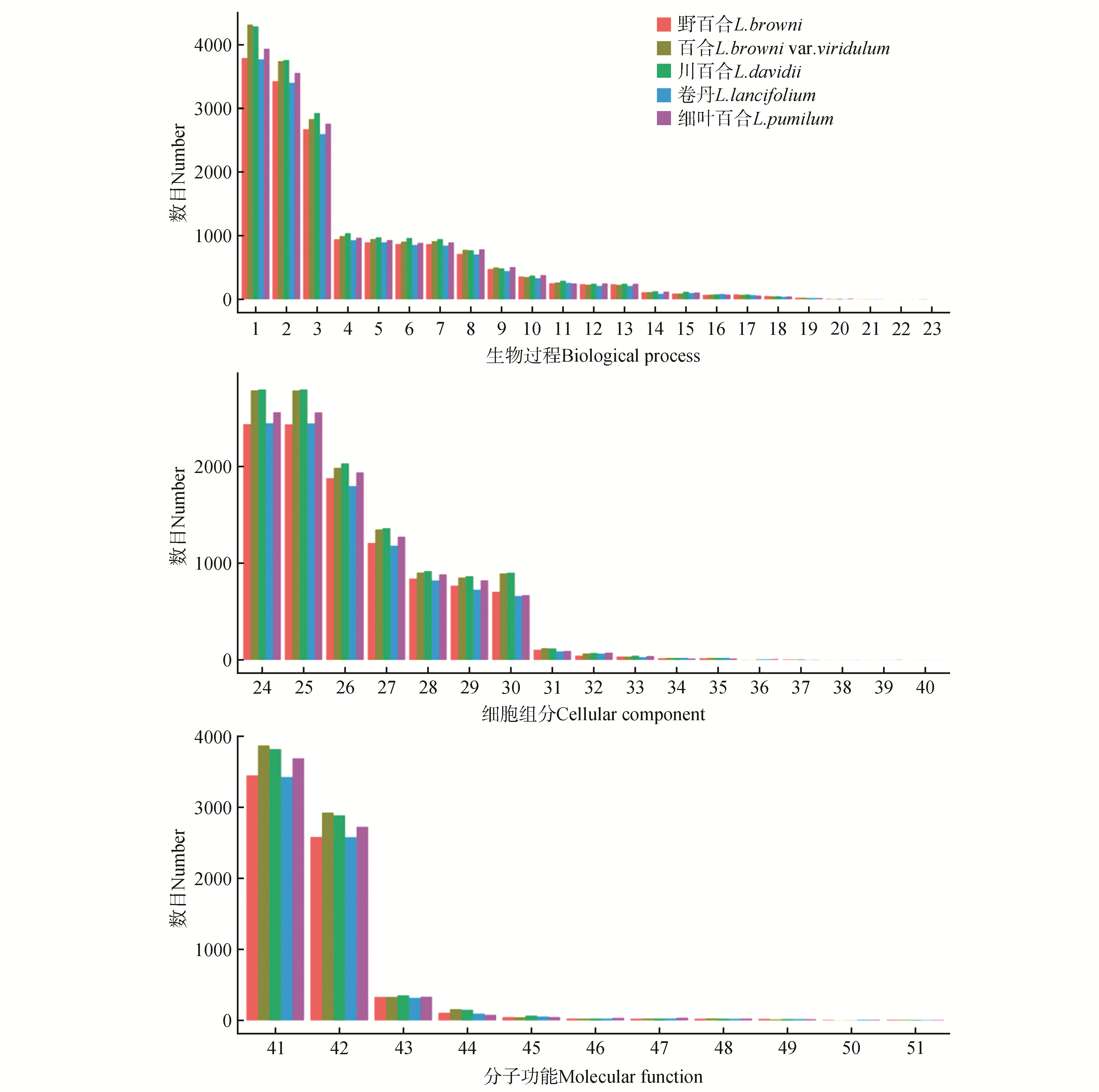
Table 3
Identified genes involved in the development of flower
基因ID Gene ID | 基因 Gene | 蛋白质 Protein |
|---|---|---|
| Unigene0017918 | MADS2 | MADS盒转录因子2 MADS-box transcription factor 2 |
| Unigene0027916 | MADS6 | MADS盒转录因子6 MADS-box transcription factor 6 |
| Unigene0004890 | MADS14 | MADS盒转录因子14 MADS-box transcription factor 14 |
| Unigene0039518 | MADS16 | MADS盒转录因子16 MADS-box transcription factor 16 |
| Unigene0040377 | AGL8 | 无性MADS盒蛋白AGL8 Agamous-like MADS-box protein AGL8 |
| Unigene0003323 | AGL15 | 无性MADS盒蛋白AGL15 Agamous-like MADS-box protein AGL15 |
| Unigene0026977 | AGL104 | 无性MADS盒蛋白AGL104 Agamous-like MADS-box protein AGL104 |
| Unigene0031138 | AGL19 | 无性MADS盒蛋白AGL19 Agamous-like MADS-box protein AGL19 |
| Unigene0025453 | AGL30 | 无性MADS盒蛋白AGL30 Agamous-like MADS-box protein AGL30 |
| Unigene0031144 | SOC1 | MADS盒蛋白SOC1 MADS-box protein SOC1 |
| Unigene0042321 | EJ2 | MADS盒蛋白EJ2 MADS-box protein EJ2 |
| Unigene0008053 | LIM15 | 减数分裂重组蛋白DMC1同源体 Meiotic recombination protein DMC1 homolog |
| Unigene0027070 | GlsA | 无性生殖细胞形成蛋白GlsA Gonidia forming protein GlsA |
Table 4
The enriched KEGG pathways of strongly positive selection unigenes
KEGG通路 KEGG pathway | 通路编号 Pathway ID | 基因注释 The annotation of gene |
|---|---|---|
糖基磷脂酰肌醇(GPI)-锚的生物合成 Glycosylphosphatidylinositol(GPI)-anchor biosynthesis | K05283 | GPI-锚定壁转移蛋白亚型4[可可] GPI-anchored wall transfer protein isoform 4[Theobroma cacao] |
萜类骨架生物合成 Terpenoid backbone biosynthesis | K06013 | CAAX异戊二烯基蛋白酶1同源物[非洲油棕] CAAX prenyl protease 1 homolog[Elaeis guineensis] |
mRNA监督途径 mRNA surveillance pathway | K12881 | RNA与输出因子结合蛋白2[涧生花烛] RNA and export factor-binding protein 2[Anthurium amnicola] |
苯丙素的生物合成 Phenylpropanoid biosynthesis | K00430 | 过氧化物酶57[巨桉] peroxidase 57[Eucalyptus grandis] |
植物-病原菌相互作用 Plant-pathogen interaction | K13457 | 疾病抗性蛋白RPM1类[枣椰树] disease resistance protein RPM1-like[Phoenix dactylifera] |
RNA运输 RNA transport | K12881 | RNA与输出因子结合蛋白2[涧生花烛] RNA and export factor-binding protein 2[Anthurium amnicola] |
剪接体 Spliceosome | K12881 | RNA与输出因子结合蛋白2[涧生花烛] RNA and export factor-binding protein 2[Anthurium amnicola] |
代谢途径 Metabolic pathways | K05283+K00430 | GPI-锚定壁转移蛋白亚型4[可可];过氧化物酶57[巨桉] GPI-anchored wall transfer protein isoform 4[Theobroma cacao];peroxidase 57[Eucalyptus grandis] |
次生代谢物的生物合成 Biosynthesis of secondary metabolites | K00430 | 过氧化物酶57[巨桉] peroxidase 57[Eucalyptus grandis] |
Supplemental table The sequence information of identified genes involved in lily flower development
基因ID Gene ID | 序列信息 Sequence information |
|---|---|
| Unigene0017918 | GCCGCGGCAAGATCGAGATCAAGCGGATCGAGAACTCCACCAACCGCCAGGTCACCTTCTCCAAGCGCCGCAACGGCATCATCAAGAAAGCCCGTGAGATCAGCGTCCTCTGTGAGGCCCAGGTCTCCGTCGTCATCTTCTCCAGCTCCGGCAAGATGTCCGAGTACTGCAGCCCCTCCACCTCTCTACCGAAGATCCTTGAGAGGTATCAGGTGAACTGCGGCAAGAAGATCTGGGATCCAAAGCATGAGCACCTGAGCGCGGAGATTGATAGGATCAAGAAGGAGAATGATAATATGCAGATCCAGCTGAGGCATCTGAAGGGAGAGGATCTGAACTCGTTGCAGCCGAAGGAGCTGATCCCAATCGAGGAGGCGCTGGAGAATGGCATCAGGGGCGTCCGGGAGAAGCAGAATGATTTTCTGAGGATGCTCAAGAAGAATGAAAGGATACTGGAAGAGGATAACAAACGGTTAACTTATATACTGCACCATCAGCAACTGGCAATGGATGAAAATATGAGGAACTTGGAATTTGCATATCATAATAAAGATGGGGATTTCGGTTCCCAGATGCCAATGGCCTTTCGTGTGCAGCCAATCCAGCCTAATTTACATGAGGACAAGTAGTATGAGCATCTCCTGTGCCTGCTATTTTATTTAGACATCGAGTATCTAAAGAAATTTATTATCTGTTATTCTACATGCTTAATGTTATTATTTGATATTCTTATCTCTTGATGTTCATGTAGTAATTTGATAGTATAATCGTAATATTGGACTTCATAGTAAAGAAGCTAG |
| Unigene0027916 | GGGTAAAAAAGAGTAAAGTTGAGATCAAATCCAACAAATCTTGCAACCCTTGTCTCTCCTCTCCTTATATTCCCTTCCTTTCTATAATTATCATCCTTTCTCCATTTCTTGAGAGAGAAGAGAGAGAGAGGGGATGGGGAGGGGAAGAGTGGAGCTGAAGAGGATTGAGAACAAGATCAACCGCCAAGTCACCTTCTCGAAACGCCGGAACGGGCTGCTGAAAAAGGCCTATGAGCTCTCGGTCCTCTGCGATGCGGAGGTCGCTCTCATCATCTTCTCCAGCAGAGGCAAGCTTTATGAGTTCGGCAGCGCAGGGACCAGTAAAACTCTTGAGCGTTACCAACGTAGTTGTTACACTTCTCAAGATACTAATGCCGTTGATCGCGAAACCCAGAGTTGGTATCAAGAAATGTCCAAATTGAAGATGAAATTTGAGTCTCTACAACGCTCCCAAAGGCATTTATTGGGAGAGGACCTCGGACCATTGAACGTGAAAGAATTACAGCAATTGGAACGCCAACTTGAATCTGCTTTGTCACAAGCTAGGCAAAGAAAGACTCAATTGATGTTGGAACAAGTGGACGAACTGCGTAAAAAGGAACGCCATCTCGGTGAAATTAACAAGCAACTAAAAAACAAGCTTGAGGCTGAGGGGTTTGACTTCCGATCTTTGCAAGTATCATGGGATACGGAGGATGTTGTTGGAGGAAATACATTTTCGCAGCATCATATGCCATCTAGTTCGATGACTATTGAACCCACTCTTCAGATGGGGTATCACCAATTTGTTCCTGCTGAAGAAGCTTCAATGCAAAGGAACACCGGTGCGGAGAATAATTTCATGCTAGGTTGGGTTCTTTGATTGAAGATTGGTTAGTACGTGTTATGTCTCGTAAAAGATCATCGGATTGTTTAACTATGTTTTCAGTTGCTTTCTTAGCTATTGTATTTCGACTTCTTTCAAATTTGCTGCTGTGGCTGATCTGCTTGTATCCGAAAGTTTTTGTTGTTGAAAATTTCATGCAGATTAATTACTGGATGGTTAGATGTATTGAATTCGGATGAATAAATTGTGTAAGGTGATGCTGTTTTTATGATGCAGCATT |
| Unigene0004890 | AAAAGTTATGCATACCAGTACATAAACCTATACATAGATATAGTTATGCCCCCTTTCTTCGAGAGGGGTAACACCATAGCAAAACCAGATCTCTGCTGCTATAAGAATGATAGTGAAACTTAGCAGAATTCCCCGTAAAATAATTATGATGGGCGGGGAAGGTGATAGCTTATCTGTTAACGTGGCGAAGCATCCATGGCGGTAGTGAGCTGCTGCCAACACGCATCAGCGGCTGTTCAGCTGCCCCCTCGTCTGCGTCGCTGCTGCTGCTGCCTCTTTCTGGGTAGATTCCAGCGTTTTGGACGGGGTGCAAATGGGGCATTAGGAAAGGAGGGGGCCTTGTCATGGGCTGTTGTCCTTGTAGCTCCCAGTGATCTTGCTGTGTCAAAGCCTCCATCCTCTGGAACTCCATGAGCTTTTTCTCCATTATAGTGTTTTGTTCCCTCAGTGATTTCTCTGTTCGCTGGAGCTCGGTAATCGAATCAAACAATAGTTGGTGCTTCCTTGATCTTACATGCTTGAAAGATAATTCGAGCTGCTGCTCTAGTTGTTGGAGTTGTTTCAGAGTCAAGTCCTCAAGCTGCTCCCCCATGAGATGTCTTTGGCTTTTCTGCAAGGTTTCAACCTTAGCCTTCAGTCTCCCGTACTCTTGACACCAGTTTCCCTGTGCTTCAGGATATGTGTCTGTCACTGCAGTTTCTGCTTGAGAATAAAGCTCGTAACGTTCGAGAATCCTTTCCATG |
| Unigene0039518 | AAGAAGATCGAAAACTCAACGAATCGGCAGGTCACTTACTCGAAGCGCCGGACTGGAATCATCAAGAAGGCGACTGAGCTCACTGTGCTCTGTGATGCCGAGGTCTCTCTCCTTATGTTCTCCAGCACCGGGAAGCTGTCAGAGTTCTGCAGCCCCTCCACAGACACGAAAAAGATCTTCGATCGCTACCAGCAGCTGTCTGGTATCAACCTCTGGAGCGCGCAATACGAGAAAATGCAAAACACTTTGAACCATCTGAGCGAGATCAACCGCAACCTTCGCAAGGAGATCAGCCAGAGGATGGGGGAGGAGCTGGATGGATTGGACATCAAGGACCTGCGCGGTCTTGAGCAAAATTTGGACGAAGCGCTCAAGCTCGTTCGTCACCGCAAGTATCATGTGATCAACACTCAGACAGAGACTTACAAGAAAAAGGTCAAAAACTCGGAAGAAGCACACAAGAACTTGCTCCGTGACCTGGTGAATCGAGAGATGAAAGATGAGAATCCAGTCTATGGTTATGTGGACGAAGACCCCAGCAACTATGATGGCGGCCTTGGTCTGGCCAATGGGGCTTCTCATCTGTACGAGTTCCGAGTCCAACCGAGCCAGCCAAACCTGCACGGGATGGGTTATGGCTCCCATGATCTCCGACTGGCTTGAAACCCTAATTCTGTAGGGTGCAGTGA |
| Unigene0040377 | GAATAACGCTCATAGCGTTCCAGAATTCTTTCCATGCTAGCGTCAGTAGAGTACTCGTACAGCTTGCCCTTGGCGGAGAATACAACAAGAGCGACCTCTGCATCGCAGAGGACAGAGATCTCATGCGCCTTCTTCAGCAGCCCCGACCGCCGCTTCGAGAAGGTGACTTGCCGGTTGATCTTGTTCTCTATCCGCTTCAGCTGCACTCGCCCCCGGCCCATCTCCAGAAAACCCTAAAACTATTGAGATGGATGAGATATCCTCAGCTTATTGAAGAGTGAGGGGGAGACAGAA |
| Unigene0003323 | CGCCAGAGGGAGTGAGTGAGACCCATAGAGGGAAATTTTGGAGGGAAAAAAATGGGGAGGAGGAAGAGGGAGATAAGGCGGATTGAGGACACCAACAGCCGCCAGGTCACTTTCTTGAAACGCGGCAGTCGGCTGCTGAAGAAGGCTTCAGAGCTCTCCGTGTTTTGCGATGCCGAGGTTGCGCTGATCGTCTTCTCACCCATGGGCAATCTCTGCGAGTTCTCCAGCTGCAGGTTAGGGTGGTATCGGTTCCAATCAAGGAGTATCCTGCTTGGCCGCCTGCAACGCGGACCAGCTGATTGGAATTTTGTTTATAATGCTGCTACTTTTAGAATAAC |
| Unigene0026977 | AGAGCTTTCTTCTCAATTACCAAAAGTATAATATTTCCTCTTGTGGTTCTAAATACAAATATTGCTCATAGATGGAGCCTATACCTCAACTCTACTTTTAGTGTATATTTAGCATATACAAATATATTTTCTAATTCTTTTATCATTATGGTCCTTACCCTATGTTCGCATTTATCTGAACAATGCTGCCTTCATAGGCAGCAGCGGTTCCTTCATACGCAGCAGCGGAGCATGATGCATTCTCGTTCCCTGGCATATGCGAGCAGTTCGCAGATGGATCCAGCTGCTCATGTGGCACCATCGATGGCAGCTCCGCTGATGCCATTGAATGCTGGATCATCGGGTAGGGACCAGTGGGGATGAGAGACGAGAGGAGCTCAGTAGAGGTGTATGCCTGGTGGTGGTGCCACATGTCGTTTTGACTGTTTATGTGACAACCATCCATGCTGGGATCCACCTGTAGCCCTGTGTTCTGGTGGATCGGGTTGTAAATCCCTTGATCCCTGAGGGACATTAGTAGATCGGACCCAACAAACATCTGATGGCCTGAGTTGGATGTTCCCTCAGTGATCCATTGGACCATCTCGTTTCCGAACGCATTGGGGATTCCTTCTTGATGAGGTTGCATGTACATCTGCATATTAGATGTAGAGGGATCGAATGTCGACAGATGATTATTTAGCAAATATTTCTTTCTCTCGTTAACTCTTGTTAGTGTCTCAATGAGGAACTTTTCACAAGTTTGCAGCTCATCCATTGATCCCATACTCAAAGGATCCGGCTCGAAACACCTTAATTGATTTTCCGAGTGTTGTAATTGTTCTTGGTATCTACTAATCTCTTGGTGGAGCTCCTCCATGTTGGAGTTACACACCCCAGGATTGGCTATTTGGGCCGCCATGTCACTCTCGCACTTTAGCTTCTTCAAGGTTCTGATTAAGTATTCTCTATTTTGGATCACTCCTCCCCGATCGTGCTCCGGGAGGCTGATGTATCGAGCGAGAACATCCTCGATCCTTCGACGGCCGGAGAAGTGGCTGAGGCGGCCGGAGGGGGAGAACATGATTAGGGCAATGTCGATGTCGCAGAGGATGGAGAGCTCGTAGGCCTTCTTGATGAGGCCATTGCGACGCTTGGAGAAGGTGACTTGGCGATTGGTGTTGTTCTCGATCTTCTTGATTTGGAGCTTTACGCGGCCCA |
| Unigene0031138 | AACATGAGATAGATATCAACTTCAACAGGCCTTCAGAAGGAAATATGCAGCAACTGCAGTGTGAAGCCGAGGGCCTCGCGAAGAGGATTCAAATCCTTGAAGATTATAAACGGAAGGTGATGGGTGAAAATCTGGAAACGTGTTCAATTGAAGAACTTCATGAATTAGAGGGTCAATTAGAGCAAAGTTTGAGCAAAATCAGACAAAAGAAGAATCACGAACAATCAGAGCAAATTGTGCAGTTAAAAGAACAGTTGTCTTCTTGCATTTTGCTCACAAAAATTCAGGAGAGGATTCTGTTGGAGAAGGAGAGGATTCTGTTGGAGAAGAATGCATCATTACAAGAAAAGGACACTGATTCTACAGAAGTTCTAGATGCTGACATGGAGGTACTCACGGAGCTGTTTATCGGGAGGCCTGGGTGCAGTAAAA |
| Unigene0025453 | TTAGTTTACATACATATCGTAACACTCAAAAATGAATCCGTTACACAAATAATCGAATCTCTATTTCTCAGATATAATGTACAACCTGAGGACAATATGCTTTATCTATCATGACACTATACAACGAAAGTTTTATATACGACGTATAACAGCTGAAATAATTTGAGAATCAGTATTGTTAAAACAGGTCCTGTATTTGTATAAGAGTAGCTACATACATCCCCACTATTAGTCCAGTGCCCGTCGTTGAATTAAATGACGATACCGGCGTATTTTCATTGACTGGAGGCGAGCCTAGGGGATGAACACTGCACAGGCTCAGGTGCTGTATGATCTACTGAATCTGGTCTGGTTGTTGGGGGTAAGGATGGTTATCAAACATGGGAACAGACGTGGAAGCCCATGTCTCGATGTTGGCATCATATTCAGGCCCAGGGGGCTCAAAACCATTAACTTGGTAATCAACCCGGCCCTCCTGTAAGCTAATTTCTGCATCGGGGTTGAACTTATTCTCGCTAAGCAAATTCATTCCATACGACTGATTAGGGTAGTGGCCTCCAAGCTGCAGTTGCAAGCATGAGTTTTGGCTCAAATCTAACTGCTCTTTGGTGTCTACCTGCTTTCCCATGGTAAAGTAACCAGGAAAGCCTTGAAGAGACCCTTCTGCGGAGCACTCAATATCTCTTTGGGAAAGCAGATTAGGGCCTTCTGGAAGCATCATGTGTGAACCATTACAGTCCTGAAGCCATTGCATATGTGACAGTTGTTGCTCGCCGTTAATTGTCAGGGGTAAATGCATACCATTCTGGAACTGGCCAGAACATTCTAATGACATAATTTGCTGCTTTCCCAGGTTTTCCTTGTGCATTCTGATTCGGTTCACAGACTCTTCAAGGGATTCCTCCATTGCTCTGATATGATCTATACTGTTGATCTTATCGATATCTTTCCAATAACTTAGTCTCTTTTCAGCGTCTGATATCTGAGCTTGCAAGAACCTTGAGTGATTTGTCAATTCCTCAATTGTTTGGGTGCTTGAACCTAGAAAATCTTGTATATTGACATCATGGTCGAGCTTCTTGAATGTCTTCTTCAGTGCTTCAAGGCTCTCCAACTTCCTCTTTGCCCTCTCCTGTGGAGTTAACTGAGCAAACTTCGAAATGACCTCCTCAATATTGCTGCGATCTCCCAGGCACATTGTTGGCTTCCCAGTCGGCGAGAACATGAGAAGGATGATATCAATGTCGCACAAAATCGATAGCTCCTTGGCCTTCTTTAGGATCCCGGCTCTCCGTTTAGAGTAAGTCACCTGCCGCGCACTGGTATTCTCCAATCTCTTGATTTTCAACTTCACCCTCCCCATCTGATCACCCTTGTGCTTAAAAGGGAATCGAGATAACCCTCGAAAAACAAAGGACCCAAATGCAATCAAGGCAACTGTCTCAAAATTCGTCCGATGTGCAACCGAATCAATAGATGGTATGAAAAAATTGCGAGAACCCAGAATTCGGAAGGGAAGAAGGACGCTGATGAAGATCAAAATAGGCAATTAGAATGAAAATTGAGGGTATCCCAATTCAGAGAATTGGACAACAAGCACGGGTTGGGAGTTCCGGAGATAGAACATCAAGAACACGATCTGT |
| Unigene0031144 | GCAATAGAGCAACCCTTGAGCGAGGGAGGGAGTGACAATCATAGAGGGAATTTTTTTGGAGGGAAAAAATGGCGAGGAGGAAGAGGGAGATAAGGCTGATTGAGGACACCAACAGCCGTCAGGTGACTTTCTCGAAGCGCGGCAGTGGGCTGCTGAAGAAGGCTTTCGAGCTCTCCGTGCTTTGCGATGTCGAGGTTGCGTTGATCGTCTTCTCACCCAAGGGCAAGCTCTGCGAGTTCTCCAGCTCCAGCATGCAAAGTACAATCGATCATTATCTGATGCATGAGAAAGATATCGACATAAGCAGGCTTTCAGAAAAAAATATGCAGCAAATGGAATGTGAATCTGCGGATCTCATGAAGAGCATTGAAATCCTTGAAGATTATAATAGGATGCGGTTGTGTGAAAATCTGGAAACATGTTCAATTGAAGAACTTCATGAAATAGAGGGTCAGTTAGAGCAAAGTTTGAGAACAATCAGACAAAAGAAGAATCACATTATATCAGAGCAAATTGCGCAGTTAAAAGAAAAGGAGAGGATTCTGTTGGAGAAGAATGCATCATTACAAGAAAAGTTCAAGGAAAAAACTCCACAACAGAAAACTGATTCTACGGAAGTTCTAGATGCTGACATGGAGGTGCTCACTGAGCTGTTTATTGGGAGGCCTGGGTGCAGCAAAACCCGAAGCATGCCAAGAGGTTGATTGAAATGCATGAGGCGCCGGGCAAGAGCCGGGAGCGGCAGATCGGACAATTCACATCGACCCCGCTTTGTCTTGACGGGGGAGGCGAACCGGACGACGGTCAGCTTTGTCTTGACGGGGGCGGCGACAGAGCGAGCGGAGGGGAAGCGGAGCACGGCGGTGATGATGGGGTTCGTGGAGGAGA |
| Unigene0042321 | TTTGAGATCACGCAACTGATCAAGTATTATCTGAGTCTTTTTCGACCTGATCTGATTTAAAGACTTCTCTAGTTGATTGTCAAGTTGTTCGAGTTCCTTGTTACTAAGTTGACCCAAGTTCTCGCCAAGGAGATTTCTACAATTCCACGGTTAAGAGTTTCATTAGCGAAATGGTTTACAACATAGATATTGCTACTACGGATTACAGATTTG |
| Unigene0008053 | GTGGTGGAATTGAGACACTACAGATCACAGAAGCCTTCGGTGAATTCAGGTCTGGAAAGACACAGATAGCTCACACCCTCTGCGTGTCAACACAGTTGCCGGTGAGCATGCATGGAGGCAACGGCAAGGTCGCTTACATCGACACAGAAGGCACATTCCGACCAGACCGCATTGTACCAATTGCAGAGAGATTTGGAATGGACGCGAGTGCAGTACTGGACAATATCATATATGCGCGGGCATACACCTACGAGCACCAGTACAACCTTCTTCTTGCTCTAGCTGCCAAGATGTCTGAAGAGCCTTTCCGGCT |
| Unigene0027070 | TCAGCCTCTCATTTCGGGGGAAAGAGGAAGCCCTAGAGCGCCGCCGCGTACCGATCCTCTCAATCGCTGCTCACAGATAAATTGAAGAGCTCTTCACCAGCTACCACCTGCTCACTTTATTTAAATTGTCAAGCCCATCGTCTGGGAGTACAATGAATGCCAAGAGTTGTCTCCTACTTACCTATTCATCTGAGGTTTTGAATGGAGTACCCTTATTCTTCGCATCTAATTGCCTCCCTGTGAAGGCTGTGAATCAAGAACCTGCTGGGCATGCGTTCCATGATGCTGCGCTTAAACTTTGCGGTTTGTTCGAAGAAGATGTAGTCACAGATGATCAAAGCGAGTCATCAGATGATCGAGGTCCAGCATTTATGGCATCATCAGATTCATATAGAAGCAAAAGCAAAAAGAAATCTGCTGGTAAAAGTGACCAACAAGACCACTATGCATTGCTAGGCTTGGGCCATCTACGTTTTCTGGCCACTGAGGAGCAGATACGTAAAAGTTACCGGGAAACTGCTTTGAAGCATCATCCTGACAAACAGGCTGCCCTTCTCCTTACTGAAAAAACAGAAGCTTCAAAGCAAGCAAAGAAGGATGAGATTGAAAACCATTTCAAAGATATTCAAGAAGCTTATGAAGTTCTAATTGATCCCGTGAGAAGAAGGGTCTATGACTCAACAGATGAGTTTGATGATGAAGTGCCGAGTGACTGTGCACCACAAGACTTCTTTAAGGTGTTTGGCCCAGCATTTATGAGGAACGGGAAATGGTCTGTAGTCCAGCCAGTTCCTTCCTTGGGGGATGATAAAACTTCACTGGAAGAGGTGGACAACTTTTACGACTTTTGGTATGCATTTAAGAGTTGGAGGGAGTTCCCACATGCAGATGAGTTCGAGCTAGAGCAATCAGAGTCTCGTGATCATAAGAGATGGATGGAAAGGCAGAATGCAAAGCTGAGAGAAAAGGCGAGGAAGGAAGAGTATGCACGTGTTCGTTCCCTTATTGATAATGCTTACAAGAGGGACCCCAGACTCCTTCGGAGAAAGGAACAGGAGAAAGCAGAGAAGCAGAGGAGAAAAGAGGCAAAGTATATGGCGAAGAAATTGCAAGAAGAAGAGGCAGCTAGAGCTGTTGAGGAAGAGCGGCTCCGCAAGGAGGAGGATGAAAAAAAAGCAGCAGAGGCTGCTGTGATTAATAAGAAGATAAAGGAAAAGGAGAAGAAGCTCTTGCGCAAGGAGAAGACTCGCCTTCGCACGTTATCAGTATCACTAGTGTCAGATTCTTTGCTTGATCTAACGGAGGATGATGTGGAAAAAACATGCAACTCTTTTGGATTGGAGCAGCTTCGTCATCTCTGTGACGGCATGGAAGGTAGGGAAGGAATTGAGAGGGCTCAACTGCTCAAAGCTGCAGTCAGTGGTGATATGTTGGAGATCTCTAAGAAGGAACCTAATGATTTGAAACCAAATGGTTCCACGAATTCGGGCCCGAAAAGTAATGGTTCTGTAACTTCAGCGAAACCAGTAATTATGATGTCCAGCTACGAAAAGAAAGAGAAGCCTTGGGGTAAGGAAGAGATAGAGTTGCTGAGAAAGGGTATCCAGAAATATCAGAAGGGGACATCTCGTAGGTGGGAAGTTATTTCAGAGTATATCGGCACTGGGAGATCGGTTGAAGAAATCCTAAAAGCCACAAAGACTGTTCTTCTCCAAAAGCCTGATTCTTCTAAAGCTTTTGACTCCTTCCTTGAGAAAAGGAAGCCGGCGAAAGCCATTGTGTCACCCCTTACGACCAGACTTGAATCAGAAGGTTCCACTGTTGAGGCAGGAGATGCATCTTCTAAATCAATACCAACACCGTCTTTGAGCAGCAGCAGTCCTGAAAAGCCAGACGGTACTCCTGTTTCCCTTCCCAATGGGGTTCCTTCTGTCCCTGAACAAGACACATGGTCTGCTACCCAAGAAAGAGCTCTCATTCAGGCGCTAAAGACTTTCCCAAAGGATGTGAATCAGCGTTGGGAACGGGTTGCTGCTGCTATTCCCGGGAAAACCATGAACCAGTGCAGAAAGAAATTTTTATCAATGAAGGAGGACTTTCGAAGCAAGAAGAGTGGTTAAGTCTTTTTTCTTTTGCAGGTAAGTTCAATATAGAACTAAAAAAAGATGACCTCTTCATCATGGTGCTGAAATTTTCCTCATTACTGGGCTTAGCTGCAGTTATATCTGGGTGCTCTTTTTCCCGCAACAGAAGTTTTTGTGATGCTTTAGGATGTAGAACTAGATGGGTATGTTTATTTAAGACACAACTTTGGTGAGATCTGACAGTTGCTACTCTTTCATATGTATGAAAGTTTTGCAAGCTCCTTGATGGGAGTTGGAAGCCCAGGCTCCTGACAAACAAATGGTTGCTGAAACATCAGACTGGAGTATAATCATCAAACCTAAAGGTATATTTATTTCTTAATGATCTCGAGAAATTTATTCATGTGATGCTGTTCAAAAGAGTTGGTGTAGCTCTATGTCGGCAATTTTGCAGTGACAATTATTTAGCCATCTGATGC |
| 1 | 李金鹏,郑春雨,刘井莉.百合属植物的研究进展[J].北方园艺,2013,(7):197-200. |
| Li J P,Zheng C Y,Liu J L.Research progress on Lilium plants[J].Northern Horticulture,2013,(7):197-200. | |
| 2 | 胡悦,杜运鹏,田翠杰,等.百合属植物化学成分及其生物活性的研究进展[J].食品科学,2018,39(15):323-332. |
| Hu Y,Du Y P,Tian C J,et al.A review of chemical components and their bioactivities from the genus Lilium[J].Food Science,2018,39(15):323-332. | |
| 3 | 杨柳慧,汪王,刘艳秋,等.细叶百合LpWRKY20基因的克隆和表达分析[J].西北林学院学报,2019,34(3):104-110. |
| Yang L H,Wang W,Liu Y Q,et al.Cloning and expression analysis of LpWRKY20 gene from Lilium pumilum[J].Journal of Northwest Forestry University,2019,34(3):104-110. | |
| 4 | 中华人民共和国药典委员会.中国药典[M].北京:中国医药科技出版社,2015:132. |
| Chinese Pharmacopeia Committee.Phamacopoeia of the People’s Republic of China[M].Beijing:China Medical Science and Technology Press,2015:132. | |
| 5 | 张卫,王嘉伦,张志杰,等.经典名方药用百合本草考证[J].中国中药杂志,2019,44(22):5007-5011. |
| Zhang W,Wang J L,Zhang Z J,et al.Herbal textual research on triditional Chinese medicine “Baihe”(Lilii Bulbus)[J].China Journal of Chinese Materia Medica,2019,44(22):5007-5011. | |
| 6 | 周佳民,宋荣,曹亮,等.不同百合(品)种生长发育特性、光合特性的比较分析及综合评价[J].中国中药杂志,2019,44(21):4581-4587. |
| Zhou J M,Song R,Cao L,et al.Comparative analysis and comprehensive evaluation for growth and photosynthetic characteristics of different lily species and varieties[J].China Journal of Chinese Materia Medica,2019,44(21):4581-4587. | |
| 7 | 苏菁华.渥丹与川百合组培快繁体系的研究[D].武汉:华中农业大学,2014. |
| Su J H.Researches on tissue culture and rapid propagation of Lilium concolor and Lilium davidii[D].Wuhan:Huazhong agricultural university,2014. | |
| 8 | Ai B,Gao Y,Zhang X L,et al.Comparative transcriptome resources of eleven primulina species,a group of “stone plants” from a biodiversity hot spot[J].Molecular ecology resources,2015,15(3):619-632. |
| 9 | 刘合霞,李博.牛耳朵与黄花牛耳朵比较转录组学分析[J].分子植物育种,2019,17(6):1853-1863. |
| Liu H X,Li B.Comparative transcriptome analysis of Primulina eburnea and Primulina lutea[J].Molecular Plant Breeding,2019,17(6):1853-1863. | |
| 10 | 赵建花.地黄属五种植物的比较转录组研究[D].西安:西北大学,2017. |
| Zhao J H.Comparative transcriptome analysis on five species of Rehmannia[D].Xi’an:Northwest University,2017. | |
| 11 | Guo W H,Jeong J,Kim Z,al Wet.Genetic diversity of Liliumtsingtauense in China and Korea revealed by ISSR markers and morphological characters[J].Biochemical Systematics and Ecology,2011,39(4-6):352-360. |
| 12 | 梅芳,李晓玲,张德春.神农架自然保护区宜昌百合染色体核型分析[J].安徽农业科学,2011,39(20):12111-12112,12115. |
| Mei F,Li X L,Zhang D C.Karyotype analysis of Lilium leucanthum(Baker) Baker in Shennongjia national nature reserve[J].Journal of Anhui Agricultural Sciences,2011,39(20):12111-12112,12115. | |
| 13 | Du Y P,He H B,Wang Z X,et al.Molecular phylogeny and genetic variation in the genus Lilium native to China based on the internal transcribed spacer sequences of nuclear ribosomal DNA[J].Journal of Plant Research,2014,127(2):249-263. |
| 14 | Li L,Stoeckert C J JR,Roos D S.OrthoMCL:identification of ortholog groups for eukaryotic genomes[J].Genome Research,2003,13(9):2178-2189. |
| 15 | Young M D,Wakefield M J,Smyth G K,et al.Gene ontology analysis for RNA-seq:accounting for selection bias[J].Genome Biology,2010,11(2):R14. |
| 16 | Yang Z H.PAML 4:phylogenetic analysis by maximum likelihood[J].Molecular Biology and Evolution,2007,24(8):1586-1591. |
| 17 | Kanehisa M,Araki M,Goto S,et al.KEGG for linking genomes to life and the environment[J].Nucleic Acids Research,2008,36(Database issue):D480-D484. |
| 18 | Grabherr M G,Haas B J,Yassour M,et al.Full-length transcriptome assembly from RNA-Seq data without a reference genome[J].Nature Biotechnology,2011,29(7):644-652. |
| 19 | Prasad K,Vijayraghavan U.Double-stranded RNA interference of a rice PI/GLO paralog,OsMADS2,uncovers its second-whorl-specific function in floral organ patterning[J].Genetics,2003,165(4):2301-2305. |
| 20 | Tzeng T Y,Chen H Y,Yang C H.Ectopic expression of carpel-specific MADS box genes from lily and lisianthus causes similar homeotic conversion of sepal and petal in Arabidopsis[J].Plant Physiology,2002,130(4):1827-1836. |
| 21 | 杨梁,王爱然,唐金富,等.百合花发育的分子生物学研究进展[J].华北农学报,2006,21(S1):81-85. |
| Yang L,Wang A R,Tang J F,et al.Research progress of molecular biology about flower development in Lily[J].Acta Agriculturae Boreali-Sinica,2006,21(S1):81-85. | |
| 22 | Jeon J S,Lee S,Jung K H,et al.Production of transgenic rice plants showing reduced heading date and plant height by ectopic expression of rice MADS-box genes[J].Molecular Breeding,2000,6(6):581-592. |
| 23 | Ohmori S,Kimizu M,Sugita M,et al.MOSAIC FLORAL ORGANS1,an AGL6-like MADS box gene,regulates floral organ identity and meristem fate in rice[J].The Plant Cell,2009,21(10):3008-3025. |
| 24 | Wang S,Huang H J,Han R,et al.Negative feedback loop between BpAP1 and BpPI/BpDEF heterodimer in Betula platyphylla × B.pendula[J].Plant Science,2019,289:110280. |
| 25 | Nagasawa N,Miyoshi M,Sano Y,et al.SUPERWOMAN1 and DROOPING LEAF genes control floral organ identity in rice[J].Development,2003,130(4):705-718. |
| 26 | Lee S,Jeon J S,An K,et al.Alteration of floral organ identity in rice through ectopic expression of OsMADS16[J].Planta,2003,217(6):904-911. |
| 27 | Wang S,Huang H J,Han R,et al.BpAP1 directly regulates BpDEF to promote male inflorescence formation in Betula platyphylla × B.pendula[J].Tree Physiology,2019,39(6):1046-1060. |
| 28 | Adamczyk B J,Fernandez D E.MIKC* MADS domain heterodimers are required for pollen maturation and tube growth in Arabidopsis[J].Plant Physiology,2009,149(4):1713-1723. |
| 29 | Bemer M,Hvan Mourik,Muiño J M,et al.FRUITFULL controls SAUR10 expression and regulates Arabidopsis growth and architecture[J].Journal of Experimental Botany,2017,68(13):3391-3403. |
| 30 | Schönrock N,Bouveret R,Leroy O,et al.Polycomb-group proteins repress the floral activator AGL19 in the FLC-independent vernalization pathway[J].Genes & development,2006,20(12):1667-1678. |
| 31 | Soyk S,Lemmon Z H,Oved M,et al.Bypassing negative epistasis on yield in tomato imposed by a domestication gene[J].Cell,2017,169(6):1142-1155.e12. |
| 32 | Nara T,Saka T,Sawado T,et al.Isolation of a LIM15/DMC1 homolog from the basidiomycete Coprinus cinereus and its expression in relation to meiotic chromosome pairing[J].Molecular and General Genetics,1999,262(4):781-789. |
| 33 | Mori T,Kuroiwa H,Higashiyama T,et al.Identification of higher plant GlsA,a putative morphogenesis factor of gametic cells[J].Biochemical and Biophysical Research Communications,2003,306(2):564-569. |
| 34 | 孟祥鹏.拟南芥抗病基因RIN4和RPM1的克隆及在大豆中的功能验证[D].长春:吉林农业大学,2018. |
| Meng X P.Cloning of arabidopsis resistance gene RIN4 and RPM1 and functional verification in soybean[D].Changchun:Jilin Agricultural University,2018. | |
| 35 | 王玉,杨雪,杨蕊菁,等.调控苯丙烷类生物合成的MYB类转录因子研究进展[J].安徽农业大学学报,2019,46(5):859-864. |
| Wang Y,Yang X,Yang R J,et al.Advances in research of MYB transcription factors in regulating phenylpropane biosynthesis[J].Journal of Anhui Agricultural University,2019,46(5):859-864. | |
| 36 | Zhao L,Zhang N,Ma P F,et al.Phylogenomic analyses of nuclear genes reveal the evolutionary relationships within the BEP clade and the evidence of positive selection in Poaceae[J].PLoS One,2013,8(5):e64642. |
| 37 | Zhang J,Xie P H,Lascoux M,et al.Rapidly evolving genes and stress adaptation of two desert poplars,Populus euphratica and P.pruinosa[J].PLoS One,2013,8(6):e66370. |
| [1] | Zhanmin ZHENG, Yubing SHANG, Guangbo ZHOU, Di XIAO, Yi LIU, Xiangling YOU. Genetic Transformation and Function Analysis of PsnHB13 and PsnHB15 of Populus simonii × Populus nigra [J]. Bulletin of Botanical Research, 2023, 43(3): 340-350. |
| [2] | Dongmei HUANG, Ying CHEN, Lu BAI, Di’an NI, Yiyang XU, Zhiguo ZHANG, Qiaoping QIN. Transcriptome Analysis of Hemerocallis fulva Leaves Respond to Low Temperature Stress [J]. Bulletin of Botanical Research, 2022, 42(3): 424-436. |
| [3] | Jingya Yu, Mingze Xia, Hao Xu, Faqi Zhang. Comparative Transcriptome Analysis of Three Artemisia Species in Qinghai Tibet Plateau [J]. Bulletin of Botanical Research, 2022, 42(2): 200-210. |
| [4] | Yun TENG, Zong-Bo QIU, Wen-Li WANG, Xue-Ru ZHANG. Transcriptome Analysis in Vascular Cambium and Xylem of Paulownia tomentosa [J]. Bulletin of Botanical Research, 2021, 41(3): 354-361. |
| [5] | Chen-Chen ZHOU, Yi-Liang LI, Zhe WANG, Fen-Cheng ZHAO, Sui-Ying ZHONG, Chang-Ming LIN, Zhi-Qiang TAN, Fu-Ming LI, Hui-Shan WU, Wen-Bing GUO, Fang-Yan LIAO, Jian-Ge WANG. Screening and Expression Analysis of Different Genes for Oleoresin Production in the Specific Period of Pinus elliottii×P.caribaea by RNA-Seq Technology [J]. Bulletin of Botanical Research, 2021, 41(3): 419-428. |
| [6] | Lin LU, Shang-Yu YANG, Wei-Dong LIU, Li-Ming LU. To Explore the Transcription Factor in Response to Low Temperature Stress in Nicotiana alata by Transcriptome Sequencing [J]. Bulletin of Botanical Research, 2021, 41(1): 119-129. |
| [7] | Li-Qiang ZHAO, Chun-Miao SHAN, Sheng-Xiang ZHANG, Yuan-Yuan SHI, Ke-Long MA, Jia-Wen WU. Identification of Key Enzyme Genes Involved in Anthocyanin Synthesis pathway in Clinopodium gracile by Transcriptome Analysis [J]. Bulletin of Botanical Research, 2020, 40(6): 886-896. |
| [8] | LIU Dan, ZENG Qin-Meng, LIU Bin, LI Yu, CHEN Shi-Pin. Transcriptome Analysis and Gene Functional Annotation in Phoebe bournei [J]. Bulletin of Botanical Research, 2020, 40(4): 613-622. |
| [9] | ZHANG Yu, XIA Ming-Ze, ZHANG Fa-Qi. Transcriptome Analysis for Medicinal Plant Anisodus tanguticus [J]. Bulletin of Botanical Research, 2020, 40(3): 458-467. |
| [10] | ZHU Li-Li, DU Qing-Xin, HE Feng, QING Jun, DU Hong-Yan. Sequencing Analysis of Transcriptome of Male Floral Bud at Two Development Stages in Eucommia ulmoides [J]. Bulletin of Botanical Research, 2020, 40(2): 284-292. |
| [11] | YE Xing-Zhuang, LIU Dan, LUO Jia-Jia, FAN Hui-Hua, ZHANG Guo-Fang, LIU Bao, CHEN Shi-Pin. Transcriptome Analysis for Rare and Endangered Plants of Semiliquidambar cathayensis [J]. Bulletin of Botanical Research, 2019, 39(2): 276-286. |
| [12] | LIU Xiong-Fang, LI Tai-Qiang, ZHANG Xu, LI Zheng-Hong, WAN You-Ming, AN Jing, LIU Xiu-Xian, MA Hong. Characteristic Analysis of Microsatellites in the Transcriptome of Phyllanthus emblica,an Important Edible and Medicinal Plant [J]. Bulletin of Botanical Research, 2019, 39(2): 294-302. |
| [13] | WANG Da-Wei, ZHOU Fan, SHEN Bing-Qi, WANG Lian-Chun. Characteristics of Microsatellite in Camellia saluenensis by High-throughput Sequencing [J]. Bulletin of Botanical Research, 2019, 39(1): 148-155. |
| [14] | LI Yan, GENGJI Zhuo-Ma, JIA Liu-Kun, WANG Zhi-Hua, CHEN Shi-Long, GAO Qing-Bo. Analysis of SSR Information in Transcriptome Sequences of Saxifraga sinomontana [J]. Bulletin of Botanical Research, 2018, 38(6): 939-947. |
| [15] | WANG Rong, CHENG Meng-Lin, LIU Hui-Na, ZHAO Da-Qiu, TAO Jun. Effects of Melatonin on Flavonoids Content and Related Gene Expression Levels of Gardenia under Dark Conditions [J]. Bulletin of Botanical Research, 2018, 38(4): 559-567. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||