Bulletin of Botanical Research ›› 2022, Vol. 42 ›› Issue (4): 613-625.doi: 10.7525/j.issn.1673-5102.2022.04.011
• Genetic and Breeding • Previous Articles Next Articles
Xueying WANG1, Ruiqi WANG1, Yang ZHANG1, Cong LIU1, Dean XIA1, Zhigang WEI2( )
)
Received:2021-05-24
Online:2022-07-20
Published:2022-07-15
Contact:
Zhigang WEI
E-mail:zhigangwei1973@163.com
About author:WANG Xueying(1996—),male,postgraduate,mainly engaged in forest tree genetics and breeding research.
Supported by:CLC Number:
Xueying WANG, Ruiqi WANG, Yang ZHANG, Cong LIU, Dean XIA, Zhigang WEI. Genome‑wide Identification and Stress Response Analysis of Cyclic Nucleotide-gated Channels(CNGC) Gene Family in Populus trichocarpa[J]. Bulletin of Botanical Research, 2022, 42(4): 613-625.
Add to citation manager EndNote|Ris|BibTeX
URL: https://bbr.nefu.edu.cn/EN/10.7525/j.issn.1673-5102.2022.04.011
Table 1
Primers for qRT-PCR of PtrCNGC gene family
基因名称 Gene name | 上游引物 Forward primer(5′→3′) | 下游引物 Reverse primer(5′→3′) |
|---|---|---|
| PtrCNGC1.1 | CAACCGAGGTGTATTGAAAGAAG | CCTGCCTACTATGAAGACGCC |
| PtrCNGC1.2 | CTCTTGAAACCACCGAGGCT | AGCAGCCCAAGTTCTCCATC |
| PtrCNGC2.1 | AAGGCTGGTGACTTCTGTGG | GTAATCGGCGAAACTGGGA |
| PtrCNGC3.1 | CGGACGAGGTGAACTTGTGAT | AGCGAGCAAACCTTGGGAT |
| PtrCNGC6.1 | GTTACAAATCCGCGTTTCGCC | TGTTGCATGACGCTCAATCG |
| PtrCNGC9.1 | CGGAAGGAGGCGACTGAC | ACAACACTTGAATCTGAGCCG |
| PtrCNGC12.1 | CTCGCAGCAGTGGAGAACAT | GGAACTCTATCTGGCACCCTT |
| PtrCNGC12.2 | CCAAGGCTGTGGAGGTTTACT | TGGCAGGCATCACGGAG |
| PtrCNGC13.1 | CTTTGCGAAATAGCATTGGAT | CAAGGTTGTGGCTACGAGATG |
| PtrCNGC14.1 | GTAGAGGAAATGCGGGTGAAG | AGCAAAGATGGCGGTTTATGT |
| PtrCNGC15.1 | GCTACATTGTTCGTGAGGGTG | CAGATTAGATGAGGAGTGGGGAT |
| PtrCNGC15.2 | GCGTTTGATTAGCAGCAGGA | CAGGCAACTTGAATGTGGGTA |
| PtrCNGC15.3 | GATTTCTGGACAGCGTTTGATTA | TTGATACCTTATGGCTCCTTGG |
| PtrCNGC17.1 | TGACTGTAGTGCCAAGGATAACC | CGGCAAACATCCCAAACTG |
| PtrCNGC17.2 | ACGCCAAGATGTGGAATGGT | GTCCTTCGGGCAGTTCTTCA |
| PtrCNGC17.3 | CGCTGTCAAACCCCGAAC | CGCACGAACGAGCAACTG |
| PtrCNGC18.1 | TCGTCAACTCCCTCAAAGTCTAC | CGTCTAACAAGGTCCAGGCAT |
| PtrCNGC18.2 | GGAAGAATGGAGACTCAAGCG | TCCATCTGTGAGAAAAAGGGC |
| PtrCNGC18.3 | TTCTCGGGTATTTGGGCG | AGCAGGATGATGTAAAGCAAGG |
| PtrActin | AGGCAGGTTTCGCAGGAGATGA | TCCATCACCAGAATCCAGCACA |
Table 2
Domain architecture of PtrCNGC gene family
亚群 Group | 基因名称 Gene name | 登录号 Gene ID | 不同蛋白数据库中的结构域 Domain architectures in different databases | ||||
|---|---|---|---|---|---|---|---|
| Pfam | SMART | CDD | PROSIT | SurperFamily | |||
| Ⅰ | PtrCNGC2.1 | Potri.002G170000 | ITP,cNMP | cNMP,TM | CAP-ED,PLN | cNMP | cAMP_bd-like,PLN |
| PtrCNGC14.1 | Potri.014G097900 | ITP,cNMP | cNMP,TM | CAP-ED,PLN | cNMP,IQ | cAMP_bd-like,PLN | |
| PtrCNGC12.1 | Potri.012G002200 | ITP,cNMP | cNMP,TM | CAP-ED,PLN | cNMP | cAMP_bd-like,PLN | |
| PtrCNGC15.1 | Potri.015G019100 | ITP,cNMP | cNMP TM | CAP-ED,PLN | cNMP | cAMP_bd-like,PLN | |
| Ⅱ | PtrCNGC18.3 | Potri.018G106100 | ITP,cNMP | cNMP,TM | CAP-ED,PLN | cNMP,IQ | cAMP_bd-like,PLN |
| PtrCNGC1.1 | Potri.001G043900 | ITP,cNMP | cNMP,TM | CAP-ED,PLN | cNMP | cAMP_bd-like,PLN | |
| PtrCNGC3.1 | Potri.003G183000 | ITP,cNMP | cNMP,TM | CAP-ED,PLN | cNMP | cAMP_bd-like,PLN | |
| Ⅲ | PtrCNGC9.1 | Potri.009G010700 | ITP,cNMP | cNMP,TM | CAP-ED,PLN | cNMP,IQ | cAMP_bd-like,PLN |
| PtrCNGC13.1 | Potri.013G108200 | ITP,cNMP | cNMP,TM | CAP-ED,PLN | cNMP | cAMP_bd-like,PLN | |
| PtrCNGC17.1 | Potri.017G067000 | ITP,cNMP | cNMP,TM | CAP-ED,PLN | cNMP | cAMP_bd-like,PLN | |
| PtrCNGC18.2 | Potri.018G097600 | ITP,cNMP | cNMP,TM | CAP-ED,PLN | cNMP | cAMP_bd-like,PLN | |
| PtrCNGC6.1 | Potri.006G271500 | ITP,cNMP | cNMP,TM | CAP-ED | cNMP | cAMP_bd-like,PLN | |
| PtrCNGC18.1 | Potri.018G009200 | ITP,cNMP | cNMP,TM | ITP,CAP-ED | cNMP | cAMP_bd-like,PLN | |
| Ⅳa | PtrCNGC12.2 | Potri.012G038700 | ITP | cNMP,TM | CAP-ED,PLN,ITP | cNMP | cAMP_bd-like,PLN |
| PtrCNGC15.2 | Potri.015G033900 | ITP,cNMP | cNMP,TM | PLN | cNMP | cAMP_bd-like,PLN | |
| PtrCNGC15.3 | Potri.015G034000 | ITP | cNMP,TM | CAP-ED,PLN,ITP | cNMP | cAMP_bd-like,PLN | |
| Ⅳb | PtrCNGC1.2 | Potri.001G407800 | ITP,cNMP | cNMP,TM | CAP-ED,PLN | cNMP | cAMP_bd-like,PLN |
| PtrCNGC17.2 | Potri.017G089800 | ITP,cNMP | cNMP,TM | CAP-ED,PLN | cNMP | cAMP_bd-like,PLN | |
| PtrCNGC17.3 | Potri.017G089900 | ITP,cNMP | cNMP,TM | CAP-ED,PLN | cNMP | cAMP_bd-like,PLN | |
Fig.1
Phylogentic Tree of CNGC gene family from Populus trichocarpa(Ptr),Arabidopsis thaliana(At),Oryza sativa(Os) and Pyrus bretchneideri(Pbr)Blue triangle,yellow triangle,red square and green circle represent Populus trichocarpa,Arabidopsis thaliana,Oryza sativa and Pyrus bretchneideri,the numbers represent the percentage of 1 000 Bootstrap replications(only above 50% showed)
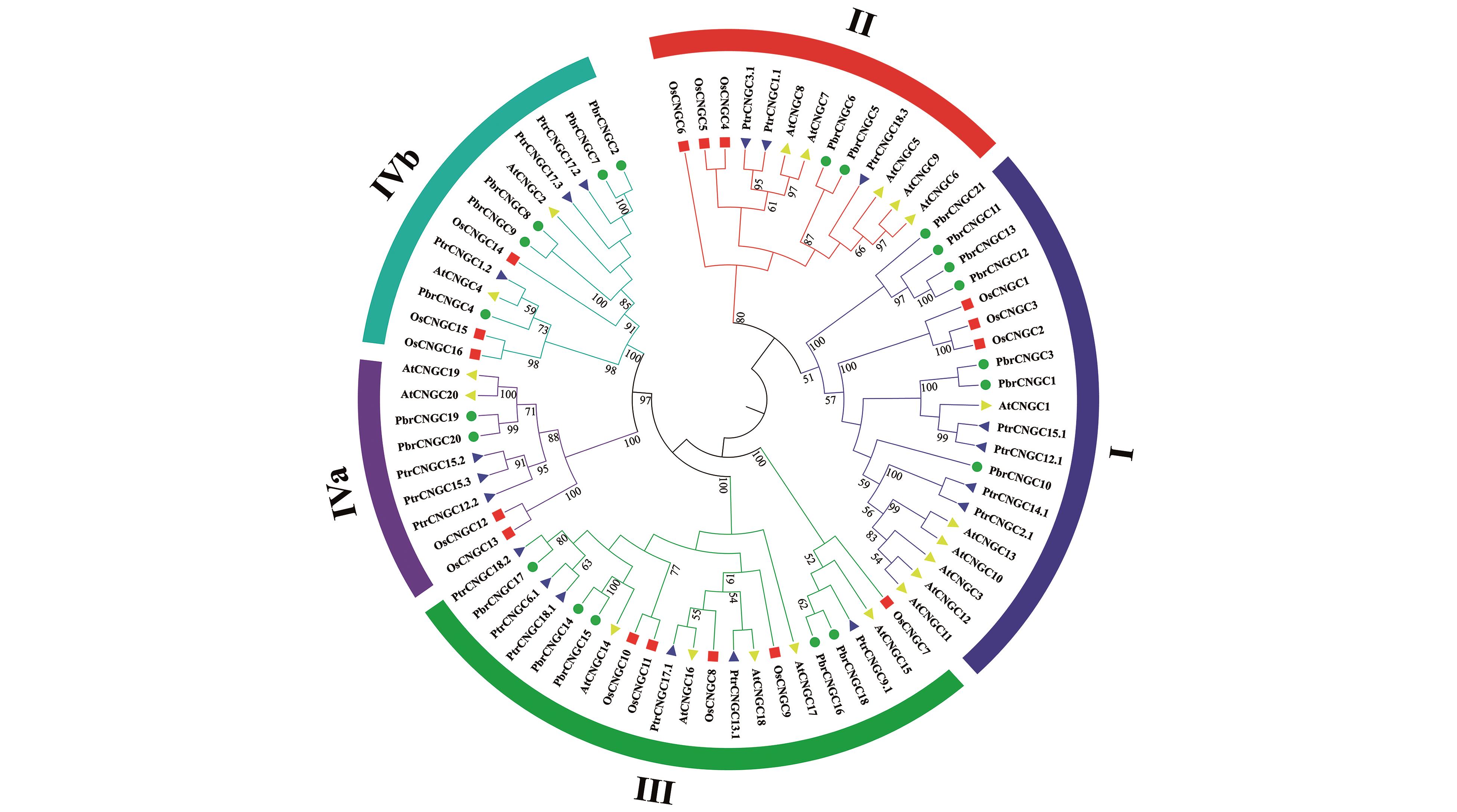
Table 3
Summary of PtrCNGC gene family
基因名称 Gene name | 登录号 Gene ID | 基因位置 Genomic location | cds长度 Length of cds /bp | 氨基酸长度 Amino acid length /aa | 等电点 pI | 分子质量 Molecular weight /kDa | 亲水性平均值 Grand average ofhydropathicity | 亚细胞定位预测(概率) Prediction of subcellular Localization(Percentage) |
|---|---|---|---|---|---|---|---|---|
| PtrCNGC1.1 | Potri.001G043900 | 3172345~3176133 | 2 238 | 745 | 9.15 | 86.026 09 | -0.266 | 细胞质(96.82%) |
| PtrCNGC1.2 | Potri.001G407800 | 43033745~43040720 | 2 055 | 684 | 8.77 | 78.532 21 | -0.119 | 细胞质(78.01%) |
| PtrCNGC2.1 | Potri.002G170000 | 12916136~12920918 | 2 187 | 728 | 9.15 | 84.103 14 | -0.193 | 细胞质(91.63%) |
| PtrCNGC3.1 | Potri.003G183000 | 18942476~18946581 | 2 220 | 739 | 8.98 | 85.236 32 | -0.193 | 细胞质(98.00%) |
| PtrCNGC6.1 | Potri.006G271500 | 27277609~27283688 | 2 076 | 691 | 7.53 | 79.603 33 | -0.226 | 细胞质(98.27%) |
| PtrCNGC9.1 | Potri.009G010700 | 1914122~1917753 | 2 238 | 745 | 9.27 | 85.463 00 | -0.131 | 细胞质(97.11%) |
| PtrCNGC12.1 | Potri.012G002200 | 206744~215632 | 2 130 | 709 | 9.03 | 82.008 80 | -0.149 | 细胞质(91.28%) |
| PtrCNGC12.2 | Potri.012G038700 | 3420369~3434999 | 2 310 | 769 | 9.23 | 88.215 84 | -0.113 | 细胞质(95.04%) |
| PtrCNGC13.1 | Potri.013G108200 | 12173969~12176818 | 2 055 | 684 | 9.32 | 79.382 80 | -0.096 | 细胞质(55.25%) |
| PtrCNGC14.1 | Potri.014G097900 | 7650920~7655559 | 2 172 | 723 | 9.45 | 83.918 28 | -0.220 | 细胞质(90.95%) |
| PtrCNGC15.1 | Potri.015G019100 | 1357777~1365061 | 2 127 | 708 | 9.11 | 81.738 42 | -0.150 | 细胞质(87.32%) |
| PtrCNGC15.2 | Potri.015G033900 | 2808538~2824047 | 2 346 | 781 | 9.42 | 89.048 90 | -0.113 | 细胞质(95.49%) |
| PtrCNGC15.3 | Potri.015G034000 | 2836520~2851182 | 2 358 | 785 | 9.53 | 89.681 54 | -0.132 | 细胞质(95.91%) |
| PtrCNGC17.1 | Potri.017G067000 | 7020102~7024372 | 2 067 | 688 | 9.16 | 79.682 99 | -0.141 | 过氧化物酶体(50.67%) |
| PtrCNGC17.2 | Potri.017G089800 | 10685838~10690733 | 2 157 | 718 | 9.49 | 82.019 61 | -0.022 | 细胞质(54.87%) |
| PtrCNGC17.3 | Potri.017G089900 | 10744470~10751288 | 2 118 | 705 | 9.55 | 81.146 94 | 0.035 | 细胞质(80.80%) |
| PtrCNGC18.1 | Potri.018G009200 | 639720~644438 | 2 163 | 720 | 9.12 | 82.685 22 | -0.148 | 细胞质(91.75%) |
| PtrCNGC18.2 | Potri.018G097600 | 12595117~12608310 | 2 184 | 727 | 9.19 | 83.793 61 | -0.236 | 细胞质(96.75%) |
| PtrCNGC18.3 | Potri.018G106100 | 13287094~13300441 | 2 205 | 734 | 9.31 | 84.049 79 | -0.217 | 细胞质(98.37%) |
Table 4
Ka/Ks ratio and homology of homologous genes
同源基因 Paralogues | 非同义替换率 Ka | 同义替换率 Ks | Ka/Ks | 同源片段长度 The length of homologous fragment /bp | 同源性 Homology /% | |
|---|---|---|---|---|---|---|
基因1 Gene 1 | 基因2 Gene 2 | |||||
| PtrCNGC1.1 | PtrCNGC3.1 | 0.056 975 | 0.220 971 | 0.257 839 | 2 026 | 90.53 |
| PtrCNGC2.1 | PtrCNGC14.1 | 0.061 054 | 0.219 205 | 0.278 523 | 1 981 | 90.37 |
| PtrCNGC6.1 | PtrCNGC18.1 | 0.094 621 | 0.306 729 | 0.308 484 | 1 355 | 89.97 |
| PtrCNGC12.1 | PtrCNGC15.1 | 0.043 307 | 0.211 946 | 0.204 330 | 1 972 | 90.53 |
| PtrCNGC12.2 | PtrCNGC15.2 | 0.115 981 | 0.274 512 | 0.422 499 | 1 572 | 85.62 |
| PtrCNGC15.3 | 0.096 435 | 0.267 658 | 0.360 292 | 1 604 | 87.36 | |
| PtrCNGC15.2 | PtrCNGC15.3 | 0.059 375 | 0.109 146 | 0.543 996 | 2 198 | 93.06 |
| 1 | YUEN C C Y, CHRISTOPHER D A.The group Ⅳ-A cyclic nucleotide-gated channels,CNGC19 and CNGC20,localize to the vacuole membrane in Arabidopsis thaliana [J].AoB Plants,2013,5:plt012. |
| 2 | 杨朝东,张霞,刘国锋,等.植物根中质外体屏障结构和生理功能研究进展[J].植物研究,2013,33(1):114-119. |
| YANG C D, ZHANG X, LIU G F,et al.Progress on the structure and physiological functions of apoplastic barriers in root[J].Bulletin of Botanical Research,2013,33(1):114-119. | |
| 3 | SCHUURINK R C, SHARTZER S F, FATH A,et al.Characterization of a calmodulin-binding transporter from the plasma membrane of barley aleurone[J].Proceedings of the National Academy of Sciences of the United States of America,1998,95(4):1944-1949. |
| 4 | NAWAZ Z, KAKAR K U, ULLAH R,et al.Genome-wide identification,evolution and expression analysis of cyclic nucleotide-gated channels in tobacco(Nicotiana tabacum L.)[J].Genomics,2019,111(2):142-158. |
| 5 | NAWAZ Z, KAKAR K U, SAAND M A,et al.Cyclic nucleotide-gated ion channel gene family in rice,identification,characterization and experimental analysis of expression response to plant hormones,biotic and abiotic stresses[J].BMC Genomics,2014,15(1):853. |
| 6 | HAO L D, QIAO X L.Genome-wide identification and analysis of the CNGC gene family in maize[J].PeerJ,2018,6(3):e5816. |
| 7 | SAAND M A, XU Y P, LI W,et al.Cyclic nucleotide gated channel gene family in tomato:genome-wide identification and functional analyses in disease resistance[J].Frontiers in Plant Science,2015,6:303. |
| 8 | CHEN J Q, YIN H, GU J P,et al.Genomic characterization,phylogenetic comparison and differential expression of the cyclic nucleotide-gated channels gene family in pear(Pyrus bretchneideri Rehd.)[J].Genomics,2015,105(1):39-52. |
| 9 | MÄSER P, THOMINE S, SCHROEDER J I,et al.Phylogenetic relationships within cation transporter families of Arabidopsis[J].Plant Physiology,2001,126(4):1646-1667. |
| 10 | KÖHLER C, MERKLE T, NEUHAUS G.Characterisation of a novel gene family of putative cyclic nucleotide- and calmodulin-regulated ion channels in Arabidopsis thaliana [J].The Plant Journal,1999,18(1):97-104. |
| 11 | ZELMAN A K, DAWE A, GEHRING C,et al.Evolutionary and structural perspectives of plant cyclic nucleotide-gated cation channels[J].Frontiers in Plant Science,2012,3:95. |
| 12 | 王文颖,柴薇薇,马清,等.植物环核苷酸门控离子通道的研究进展[J].植物生理学报,2015,51(11):1799-1808. |
| WANG W Y, CHAI W W, MA Q,et al.Research advances in cyclic nucleotide-gated channels in plant[J].Plant Physiology Journal,2015,51(11):1799-1808. | |
| 13 | 刘海娇,杜立群,林金星,等.植物环核苷酸门控离子通道及其功能研究进展[J].植物学报,2015,50(6):779-789. |
| LIU H J, DU L Q, LIN J X,et al.Recent advances in cyclic nucleotide-gated ion channels with their functions in plants[J].Bulletin of Botany,2015,50(6):779-789. | |
| 14 | CHANG F, YAN A, ZHAO L N,et al.A putative calcium-permeable cyclic nucleotide-gated channel,CNGC18,regulates polarized pollen tube growth[J].Journal of Integrative Plant Biology,2007,49(8):1261-1270. |
| 15 | 许扬.水稻控制花粉管生长基因OsCNGC13的图位克隆及功能分析和水稻卷叶基因OsZHD1的功能研究[D].南京:南京农业大学,2016. |
| XU Y.Map-based cloning and functional analysis of OsCNGC13 gene controling pollen tube growth and functional analysis of abaxially cured leaf gene OsZHD1 in rice(Oryza sativa L.)[D].Nanjing:Nanjing Agricultural University,2016. | |
| 16 | 董紫怡.拟南芥CNGC2蛋白的胞吞及其在抗病中的作用[D].北京:北京林业大学,2019. |
| DONG Z Y.Endocytosis of CNGC2 and its role in disease resistance in Arabidopsis [D].Beijing:Beijing Forestry University,2019. | |
| 17 | 林世锋,王仁刚,余婧,等.一个受青枯病菌诱导的烟草功能基因NtCNGC1的克隆与表达分析[J].中国烟草学报,2018,24(6):86-92. |
| LIN S F, WANG R G, YU J,et al.Cloning and expression analysis of a tobacco functional gene NtCNGC1 induced by Ralstonia solanacearum [J].Acta Tabacaria Sinica,2018,24(6):86-92. | |
| 18 | 段琼,王晓宇,霍红雁,等.蓖麻环核苷酸门控离子通道RcCNGC2克隆与盐胁迫下表达分析[J].华北农学报,2020,35(4):79-86. |
| DUAN Q, WANG X Y, HUO H Y,et al.Cloning and characterization of RcCNGC2 gene of Ricinus communis L.cyclic nucleotide-gated channel expression analysis under salt stress[J].Acta Agriculturae Boreali-Sinica,2020,35(4):79-86. | |
| 19 | TUSKAN G A, DIFAZIO S, JANSSON S,et al.The genome of black cottonwood,Populus trichocarpa(Torr.& Gray)[J].Science,2006,313(5793):1596-1604. |
| 20 | HALL T A.BioEdit:a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT[J].Nuclc Acids Symposium Series,1999,41:95-98. |
| 21 | CHEN C J, CHEN H, ZHANG Y,et al.TBtools:an integrative toolkit developed for interactive analyses of big biological data[J].Molecular Plant,2020,13(8):1194-1202. |
| 22 | TAMURA K, STECHER G, PETERSON D,et al.MEGA6:molecular evolutionary genetics analysis version 6.0[J].Molecular Biology and Evolution,2013,30(12):2725-2729. |
| 23 | BLANC G, WOLFE K H.Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes[J].The Plant Cell,2004,16(7):1667-1678. |
| 24 | WANG L Z, HU W, SUN J T,et al.Genome-wide analysis of SnRK gene family in Brachypodium distachyon and functional characterization of BdSnRK2.9 [J].Plant Science,2015,237:33-45. |
| 25 | LYNCH M, CONERY J S.The evolutionary fate and consequences of duplicate genes[J].Science,2000,290(5494):1151-1155. |
| 26 | LESCOT M, DÉHAIS P, THIJS G,et al.PlantCARE,a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J].Nucleic Acids Research,2002,30(1):325-327. |
| 27 | 陆业磊,邓为,王震,等.高粱SPL基因家族的鉴定及表达分析[J].生物资源,2020,42(4):444-453. |
| LU Y L, DENG W, WANG Z,et al.Identification and expression analysis of SPL gene family in Sorghum bicolor L.[J].Biotic Resources,2020,42(4):444-453. | |
| 28 | KONG H Z, LANDHERR L L, FROHLICH M W,et al.Patterns of gene duplication in the plant SKP1 gene family in angiosperms:evidence for multiple mechanisms of rapid gene birth[J].The Plant Journal,2007,50(5):873-885. |
| 29 | HOLUB E B.The arms race is ancient history in Arabidopsis,the wildflower[J].Nature Reviews Genetics,2001,2(7):516-527. |
| 30 | LI Q Q, YANG S Q, REN J,et al.Genome-wide identification and functional analysis of the cyclic nucleotide-gated channel gene family in Chinese cabbage[J].3 Biotech,2019,9(3):114. |
| 31 | ZHU L C, ZHANG Y, ZHANG W,et al.Patterns of exon-intron architecture variation of genes in eukaryotic genomes[J].BMC Genomics,2009,10(1):47. |
| [1] | Anying HUANG, Dean XIA, Yang ZHANG, Dongchen NA, Qing YAN, Zhigang WEI. Cloning and Drought Tolerance Expression Analysis of PtrWRKY51 Gene in Populus trichocarpa [J]. Bulletin of Botanical Research, 2022, 42(6): 1005-1013. |
| [2] | He CHENG, Shuanghui TIAN, Yang ZHANG, Cong LIU, De’an XIA, Zhigang WEI. Genome-wide Identification and Expression Analysis of nsLTP Gene Family in Populus trichocarpa [J]. Bulletin of Botanical Research, 2022, 42(3): 412-423. |
| [3] | Shuang-Hui TIAN, He CHENG, Yang ZHANG, Cong LIU, De-An XIA, Zhi-Gang WEI. Genome-wide Identification and Expressional Analysis of Carotenoid Cleavage Dioxygenases(CCD) Gene Family in Populus trichocarpa under Drought and Salt Stress [J]. Bulletin of Botanical Research, 2021, 41(6): 993-1005. |
| [4] | Meng-Xuan REN, Yang ZHANG, Shuang WANG, Rui-Qi WANG, Cong LIU, Zhi-Gang WEI. Genome-wide Identification and Expression Analysis GATA Family of Populus trichocarpa [J]. Bulletin of Botanical Research, 2021, 41(1): 107-118. |
| [5] | Bing LI, Yu-Xiang CHENG. Analysis of SHMT Gene Family and Production of PtrSHMT9 Mutant in Populus trichocarpa [J]. Bulletin of Botanical Research, 2020, 40(6): 906-912. |
| [6] | DONG Shi-Wei, YANG Yu-Ning, WANG Nai-Rui, ZHANG Han-Guo, LI Shu-Juan. Gene Cloning and Stress Response Analysis of Natural Disorder Protein in Populus trichocarpa [J]. Bulletin of Botanical Research, 2020, 40(4): 575-582. |
| [7] | LI Ya-Bo, Lü Jia-Xin, TAN Bing, GAO Cai-Qiu. Cloning and expression analysis of 5 ZFP genes from Poplus trichocarpa [J]. Bulletin of Botanical Research, 2020, 40(2): 243-250. |
| [8] | WANG Shuang, ZHANG Yang, REN Meng-Xuan, LIU Ying-Ying, WEI Zhi-Gang. Genome-Wide Analysis of the WOX Family Reveals Their Involvement in Stem Growth of Populus trichocarpa [J]. Bulletin of Botanical Research, 2019, 39(4): 568-577. |
| [9] | WANG Jia-Qi, ZHANG Xi, LI Li. Bioinformatic and Expression Analysis of HD-Zip Family Gene in Betula platyphylla [J]. Bulletin of Botanical Research, 2018, 38(6): 931-938. |
| [10] | WEI Zhen, CHENG Yu-Xiang, XIA De-An. Cas9/gRNA-mediated Targeted PtrFLA31/34 Gene Mutation in Populus [J]. Bulletin of Botanical Research, 2018, 38(5): 741-747. |
| [11] | GONG Dao-Yong, HU Shang-Lian, CAO Ying, LU Xue-Qin, ZHANG Qing-Bo. Cloning and Bioinformatics Analysis of Two bZIP Genes of Bambusa emeiensis and Their Induced Expression under Abiotic Stresses [J]. Bulletin of Botanical Research, 2018, 38(2): 268-277. |
| [12] | ZHANG Xue-Mei, YAO Wen-Jing, ZHAO Kai, JIANG Ting-Bo, ZHOU Bo-Ru. Bioinformatics and Response to Salt Stress Analysis of the HD-Zip Transcription Factor Family in Populus simonii×P.nigra [J]. Bulletin of Botanical Research, 2017, 37(5): 715-721. |
| [13] | FAN A-Qi;;MAO Juan;;LIU Si-Yan;;ZHANG Jun-Lian;;WANG Di;*;BAI Jiang-Ping;. Cloning and Bioinformatic Analysis of StSnRK2.2 Gene in Solanum tuberosum [J]. Bulletin of Botanical Research, 2013, 33(3): 294-301. |
| [14] | LU Yu-Jian;LIU Heng;LI Yu-Xi;LIANG Da-Wei;ZHAO Peng;JIA Peng-Fei;NIU Song-Zhao*. Preliminary Study on the Subcellular Localization of Arabidopsis thaliana AtZW10 [J]. Bulletin of Botanical Research, 2013, 33(2): 174-180. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||