Bulletin of Botanical Research ›› 2022, Vol. 42 ›› Issue (4): 602-612.doi: 10.7525/j.issn.1673-5102.2022.04.010
• Genetic and Breeding • Previous Articles Next Articles
Yu ZHANG1,2, Xu SU1,2,3, Yuping LIU1,2( ), Tao LIU1,2, Changyuan ZHENG1,2, Dandan SU1,2, Yanan WANG1,2, Ting LÜ1,2
), Tao LIU1,2, Changyuan ZHENG1,2, Dandan SU1,2, Yanan WANG1,2, Ting LÜ1,2
Received:2021-07-21
Online:2022-07-20
Published:2022-07-15
Contact:
Yuping LIU
E-mail:lyp8527970@126.com
About author:ZHANG Yu(1998—),female,postgraduate,mainly engaged in the study of genetic diversity and systematic evolution of alpine plants.
Supported by:CLC Number:
Yu ZHANG, Xu SU, Yuping LIU, Tao LIU, Changyuan ZHENG, Dandan SU, Yanan WANG, Ting LÜ. Characteristics of Complete Chloroplast Genome and Phylogenetic Analysis of Rhodiola himalensis (Crassulaceae)[J]. Bulletin of Botanical Research, 2022, 42(4): 602-612.
Add to citation manager EndNote|Ris|BibTeX
URL: https://bbr.nefu.edu.cn/EN/10.7525/j.issn.1673-5102.2022.04.010

Fig.1
Circularized map of the chloroplast genome of R. himalensisThe species name and specific information regarding the genome(length and the number of genes) are depicted in the center of the plot;Genes on the outside and inside of the circle are transcribed in clockwise and counterclockwise directions,respectively;Genes belonging to different functional groups are color coded;The gene names and their optional codon usage bias are labeled on the outermost layer;The SSC,LSC and inverted repeat regions(IRA and IRB) are indicated
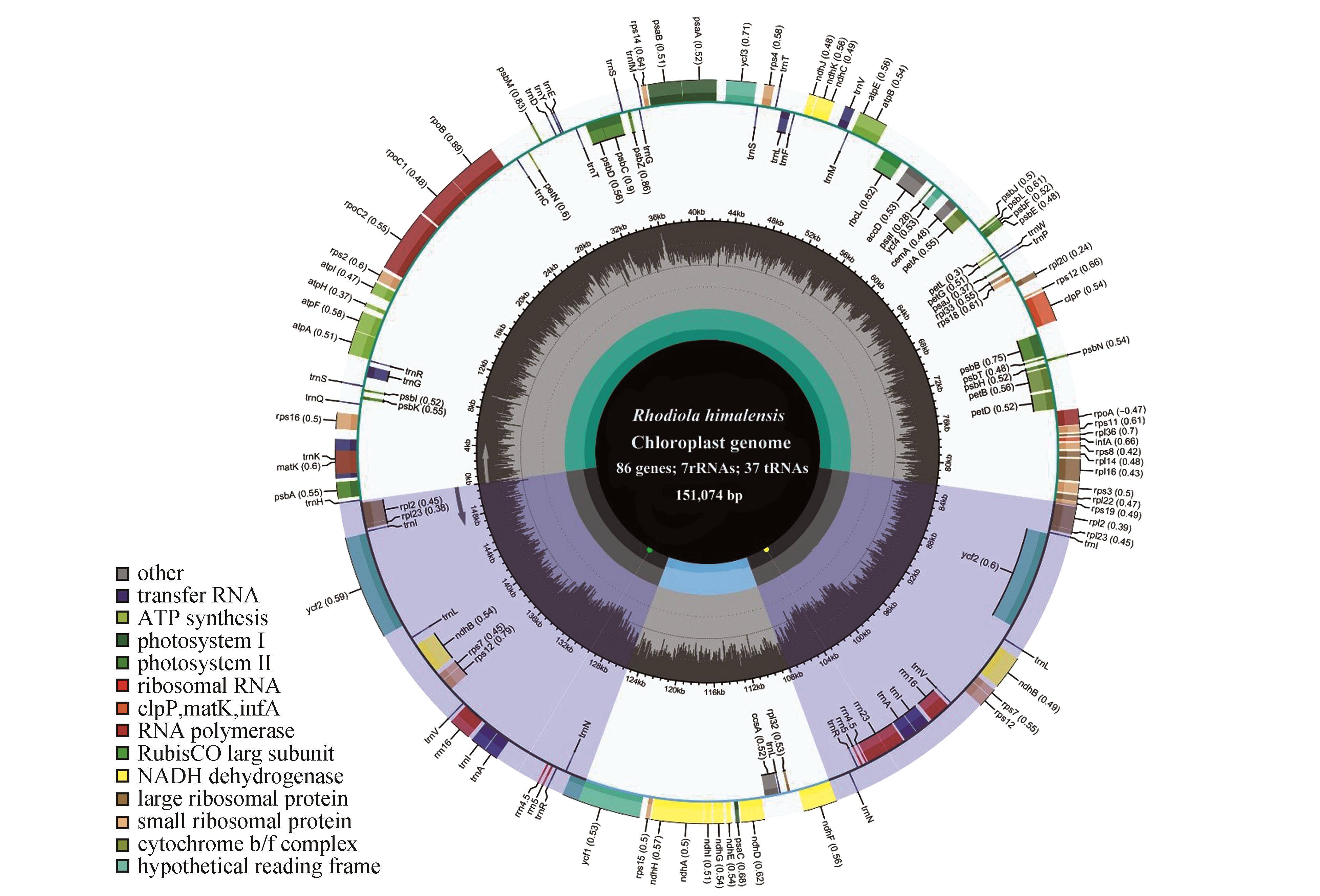
Table 2
Gene annotation of the chloroplast genome of R. himalensis
基因类别 Gene category | 基因分组 Gene group | 基因名称 Gene name |
|---|---|---|
光合作用基因 Genes for photosynthesis | 光系统 Ⅰ 亚基 Subunits of photosystem Ⅰ | psaA、psaB、psaC、psaI、psaJ |
光系统 Ⅱ 亚基 Subunits of photosystem Ⅱ | psbA、psbB、psbC、psbD、psbE、psbF、psbH、psbI、psbJ、 psbK、psbL、psbM、psbN、psbT、psbZ | |
细胞色素b/f复合体亚基 Subunit of cytochrome b/f complex | PetA、petBa 、petDa 、petG、petL、petN | |
ATP合成酶亚基 Subunits of ATP synthase | atpA、atpB、atpE、atpFa、atpH、atpI | |
NADH脱氢酶亚基 Subunits of NADH dehydrogenase | ndhAa、ndhBac、ndhC、ndhD、ndhE、ndhF、ndhG、 ndhH、ndhI、ndhG、ndhK | |
二磷酸核酮糖羧化酶大亚基 Large subunit of Rubisco | rbcL | |
ATP依赖蛋白酶亚基P基因 ATP-dependent protease subunit P | clpPb | |
自我复制基因 Self replication | DNA依赖性RNA聚合酶 DNA dependent RNA polymerase | rpoA、rpoB、rpoC1a、rpoC2 |
核糖体小亚基 Small subunit of ribosome | rps2、rps3、rps4、rps7c、rps8、rps11、rps12d*、rps14、 rps15、rps16a、rps18、rps19 | |
核糖体大亚基 Large subunit of ribosome | rpl2ac、rpl14、rpl16、rpl20、rpl22、rpl23c、rpl32、 rpl33、rpl36 | |
转运RNA Transfer RNA gene | trnA-UGCac、trnC-GCA、trnD-GUC、trnE-UUC、trnF-GAA、trnG-GCC、trnG-UCC、trnH-GUG、trnI-CAUc、trnI-GAUac、trnK-UUUa、trnL-CAAc、trnL-UAAa、trnL-UAG、trnM-CAU、trnN-GUUc、trnP-UGG、trnQ-UUG、trnR-ACGc、trnR-UCU、trnS-GCU、trnS-GGA、trnS-UGA、trnT-GGU、trnT-UGU、trnV-GACc、trnV-UACa、trnW-CCA、trnY-GUA、trnfM-CAU | |
核糖体RNA Ribosomal RNA gene | rrn4.5c、rrn5c、rrn16c、rrn23 | |
翻译起始因子 Translation initiation factor | infA | |
其他基因 Other genes | 成熟酶 Maturase | matK |
包膜蛋白 Envelop membrane protein | cemA | |
c型细胞色素合成基因 c-type cytochrome synthesis gene | ccsA | |
乙酰辅酶A羧化酶亚基 Submit of acetyl-CoA-carboxylase | accD | |
未知功能基因 Genes of unknown function | 开放阅读框 Conserved open reading frame | ycf1、ycf2c、ycf3b、ycf4 |
Table 3
RSCU analysis of protein coding region in R. himalensis
氨基酸 Amino acid | 密码子 Codon | 数量 Number | RSCU | 使用频率 Ratio /% | 氨基酸 Amino acid | 密码子 Codon | 数量 Number | RSCU | 使用频率 Ratio /% |
|---|---|---|---|---|---|---|---|---|---|
| Phe | UUU | 956 | 1.33 | 5.62 | Ala | GCU | 603 | 1.79 | 5.27 |
| UUC | 477 | 0.67 | GCC | 202 | 0.60 | ||||
| Leu | UUA | 869 | 1.92 | 10.67 | GCA | 401 | 1.19 | ||
| UUG | 531 | 1.17 | GCG | 139 | 0.41 | ||||
| CUU | 591 | 1.30 | TER* | UAA | 39 | 1.70 | 0.27 | ||
| CUC | 171 | 0.38 | UAG | 16 | 0.70 | ||||
| CUA | 390 | 0.86 | UGA | 14 | 0.61 | ||||
| CUG | 169 | 0.37 | His | CAU | 484 | 1.53 | 2.48 | ||
| Ile | AUU | 1 043 | 1.46 | 8.43 | CAC | 149 | 0.47 | ||
| AUC | 425 | 0.59 | Gln | CAA | 696 | 1.54 | 3.55 | ||
| AUA | 682 | 0.95 | CAG | 210 | 0.46 | ||||
| Met | AUG | 613 | 1.00 | 2.40 | Asn | AAU | 937 | 1.51 | 4.86 |
| Val | GUU | 520 | 1.51 | 5.39 | AAC | 302 | 0.49 | ||
| GUC | 162 | 0.47 | Lys | AAA | 1 044 | 1.51 | 5.41 | ||
| GUA | 505 | 1.47 | AAG | 335 | 0.49 | ||||
| GUG | 187 | 0.54 | Asp | GAU | 835 | 1.59 | 4.11 | ||
| Ser | UCU | 559 | 1.68 | 7.81 | GAC | 214 | 0.41 | ||
| UCC | 316 | 0.95 | Glu | GAA | 1 014 | 1.51 | 5.27 | ||
| UCA | 402 | 1.21 | GAG | 330 | 0.49 | ||||
| UCG | 193 | 0.58 | Cys | UGU | 211 | 1.45 | 1.14 | ||
| AGU | 403 | 1.21 | UGC | 81 | 0.55 | ||||
| AGC | 119 | 0.36 | Trp | UGG | 450 | 1.00 | 1.76 | ||
| Pro | CCU | 403 | 1.53 | 4.13 | Arg | CGU | 344 | 1.37 | 5.92 |
| CCC | 212 | 0.81 | CGC | 91 | 0.36 | ||||
| CCA | 290 | 1.10 | CGA | 355 | 1.41 | ||||
| CCG | 148 | 0.56 | CGG | 110 | 0.44 | ||||
| Thr | ACU | 549 | 1.71 | 5.02 | AGA | 447 | 1.77 | ||
| ACC | 212 | 0.66 | AGG | 164 | 0.65 | ||||
| ACA | 386 | 1.20 | Gly | GGU | 579 | 1.33 | 6.84 | ||
| ACG | 135 | 0.42 | GGC | 162 | 0.37 | ||||
| Tyr | UAU | 762 | 1.64 | 3.65 | GGA | 699 | 1.60 | ||
| UAC | 170 | 0.36 | GGG | 306 | 0.70 |
Table 5
SSR information of the chloroplast genome in R. himalensis
编号 No. | SSR类型 SSR type | SSR | 大小 Size | 起始 Start | 终止 End | 位置 Location |
|---|---|---|---|---|---|---|
| 1 | P1 | T(11) | 11 | 2 159 | 2 169 | CDS(matK) |
| 2 | P1 | T(10) | 10 | 3 762 | 3 771 | Intron(trnK-UUU) |
| 3 | P1 | (A)10 | 10 | 5 231 | 5 240 | Intron(rps16) |
| 4 | P1 | A(11) | 11 | 7 536 | 7 546 | IGS |
| 5 | P1 | (T)12 | 12 | 8 161 | 8 172 | IGS |
| 6 | P1 | (T)15 | 15 | 9 137 | 9 151 | Intron(trnG-GCC) |
| 7 | C | (TA)7ttatatattaatatatttctatttaatatatatagatt(TA)12 | 76 | 9 369 | 9 444 | IGS |
| 8 | P1 | (T)10 | 10 | 12 061 | 12 070 | CDS(atpF) |
| 9 | P1 | (A)12 | 12 | 15 624 | 15 635 | IGS |
| 10 | P1 | (T)11 | 11 | 17 841 | 17 851 | CDS(rpoC2) |
| 11 | P1 | (T)11 | 11 | 22 005 | 22 015 | CDS(rpoC1) |
| 12 | P1 | (T)10 | 10 | 26 932 | 26 941 | IGS |
| 13 | P1 | (A)14 | 14 | 27 103 | 27 116 | IGS |
| 14 | P1 | (T)11 | 11 | 29 349 | 29 359 | IGS |
| 15 | P1 | (A)10 | 10 | 31 963 | 31 972 | IGS |
| 16 | P1 | (T)12 | 12 | 32 262 | 32 273 | IGS |
| 17 | P1 | (A)11 | 11 | 36 504 | 36 514 | IGS |
| 18 | P1 | (A)12 | 12 | 57 371 | 57 382 | CDS(accD) |
| 19 | P2 | (TA)6 | 12 | 57 830 | 57 841 | IGS |
| 20 | P1 | (A)10 | 10 | 58 230 | 58 239 | IGS |
| 21 | P1 | (T)11 | 11 | 59 093 | 59 103 | IGS |
| 22 | P1 | (A)11 | 11 | 61 422 | 61 432 | IGS |
| 23 | P1 | (T)11 | 11 | 64 766 | 64 776 | IGS |
| 24 | P1 | (T)10 | 10 | 65 122 | 65 131 | IGS |
| 25 | P1 | (T)11 | 11 | 65 794 | 65 804 | IGS |
| 26 | P1 | (A)10 | 10 | 73 561 | 73 570 | Intron(petB) |
| 27 | P1 | (A)10 | 10 | 79 265 | 79 274 | IGS |
| 28 | P1 | (T)12 | 12 | 80 743 | 80 754 | Intron(rpl16) |
| 29 | P1 | (T)12 | 12 | 81 664 | 81 675 | IGS |
| 30 | P1 | (T)10 | 10 | 100 712 | 100 721 | Intron(trnI-GAU) |
| 31 | P1 | (A)10 | 10 | 108 440 | 108 449 | CDS(ndhF) |
| 32 | P1 | (T)12 | 12 | 108 910 | 108 921 | CDS(ndhF) |
| 33 | P1 | (A)10 | 10 | 112 719 | 112 728 | IGS |
| 34 | P1 | (A)10 | 10 | 114 523 | 114 532 | IGS |
| 35 | P1 | (T)10 | 10 | 116 247 | 116 256 | CDS(ndhG) |
| 36 | P2 | (AT)7 | 14 | 117 832 | 117 845 | Intron(ndhA) |
| 37 | P1 | (A)10 | 10 | 122 439 | 122 448 | CDS(ycf1) |
| 38 | C | (T)14gattcaaaatcaaaaaaaagttgtcccttgatccttattatattaataactaggacgatagcaatgtatattctttc(A)11 | 102 | 123 070 | 123 171 | CDS(ycf1) |
| 39 | P1 | (A)11 | 11 | 123 629 | 123 639 | CDS(ycf1) |
| 40 | P1 | (T)10 | 10 | 123 763 | 123 772 | CDS(ycf1 |
| 41 | P1 | (A)11 | 11 | 125 092 | 125 102 | CDS(ycf1) |
| 42 | P1 | (A)10 | 10 | 132 664 | 132 673 | Intron(trnI-GAU) |
| 1 | 宋菊,龙月红,林丽梅,等.五加科植物叶绿体基因组结构与进化分析[J].中草药,2017,48(24):5070-5075. |
| SONG J, LONG Y H, LIN L M,et al.Analysis on structure and phylogeny of chloroplast genomes in Araliaceae species[J].Chinese Traditional and Herbal Drugs,2017,48(24):5070-5075. | |
| 2 | JANSEN R K, RAUBESON L A, BOORE J L,et al.Methods for obtaining and analyzing whole chloroplast genome sequences[J].Methods in Enzymology,2005,395:348-384. |
| 3 | LESEBERG C H, DUVALL M R.The complete chloroplast genome of Coix lacryma-jobi and a comparative molecular evolutionary analysis of plastomes in cereals[J].Journal of Molecular Evolution,2009,69(4):311-318. |
| 4 | 焦凯丽,郑凯欣,朱宇佳,等.茄科植物叶绿体基因组研究进展[J].杭州师范大学学报(自然科学版),2019,18(2):160-167. |
| JIAO K L, ZHENG K X, ZHU Y J,et al.Advances in chloroplast genome research of Solanaceae plants[J].Journal of Hangzhou Normal University(Natural Science Edition),2019,18(2):160-167. | |
| 5 | GUI L J, JIANG S F, XIE D F,et al.Analysis of complete chloroplast genomes of Curcuma and the contribution to phylogeny and adaptive evolution[J].Gene,2020,732:144355. |
| 6 | ZENG S Y, ZHAO J H, HAN K,et al.Complete chloroplast genome sequences of Rehmannia chingii,an endemic and endangered herb[J].Conservation Genetics Resources,2016,8(4):407-409. |
| 7 | RAMAN G, PARK K T, KIM J H,et al.Characteristics of the completed chloroplast genome sequence of Xanthium spinosum:comparative analyses,identification of mutational hotspots and phylogenetic implications[J].BMC Genomics,2020,21(1):855. |
| 8 | XING Y P, XU L, CHEN S Y,et al.Comparative analysis of complete chloroplast genomes sequences of Arctium lappa and A.tomentosum [J].Biologia Plantarum,2019,63(1):565-574. |
| 9 | ZHAO D N, ZHANG J Q.Characterization of the complete chloroplast genome of the traditional medicinal plants Rhodiola rosea(Saxifragales:Crassulaceae)[J].Mitochondrial DNA Part B,2018,3(2):753-754. |
| 10 | ZHAO K H, XU Y J, LU Y Z,et al.The complete chloroplast genome sequence of Rhodiola kirilowii(Crassulaceae),a precious Tibetan drug in China[J].Mitochondrial DNA Part B,2020,5(3):3128-3129. |
| 11 | 中国科学院中国植物志编辑委员会.中国植物志:第40卷[M].北京:科学出版社,1998:80. |
| Editorial Committee of Flora of China, Chinese Academy of Sciences.Flora of China:Vol.40[M].Beijing:Science Press,1998:80. | |
| 12 | 刘青.西藏高原红景天遗传多样性与质量分析比较[D].北京:北京中医药大学,2016. |
| LIU Q.Genetic diversity and quality comparison of Rhodiola in Tibet Plateau[D].Beijing:Beijing University of Chinese Medicine,2016. | |
| 13 | 李雪彤,吴委林,权伍荣.红景天属药用植物研究进展[J].延边大学农学学报,2018,40(4):83-90. |
| LI X T, WU W L, QUAN W R.Research advances on medicinal plants from Rhodioal L. [J].Journal of Agricultural Science Yanbian University,2018,40(4):83-90. | |
| 14 | 崔艳梅,娄安如,赵长琦.红景天属植物化学成分及药理作用研究进展[J].北京师范大学学报(自然科学版),2008,44(3):328-333. |
| CUI Y M, LOU A R, ZHAO C Q.Phytochemical components and their pharmacological action of Rhodiola L. [J].Journal of Beijing Normal University(Natural Science),2008,44(3):328-333. | |
| 15 | 关伟,杨长青 .高山红景天提取物对小鼠不同抑郁模型的影响[J].中国民康医学,2012,24(21):2602-2604. |
| GUAN W, YANG C Q.Effects of Rhodiola sachalinensis extract on different depression models in mice[J].Medical Journal of Chinese People’s Health,2012,24(21):2602-2604. | |
| 16 | 陈海娟.青海红景天属药用植物资源研究[D].沈阳:沈阳药科大学,2009. |
| CHEN H J.Study on medicinal plant resources of Rhodiola in Qinghai[J].Shenyang:Shenyang Pharmaceutical University,2009. | |
| 17 | 邱林刚,陈金瑞,蒋思平,等.喜马红景天的化学成分[J].云南植物研究,1989(2):219-222. |
| QIU L G, CHEN J R, JIANG S P,et al.The chemical constituents of Rhodiola himalensis [J].Acta Botanica Yunnanica,1989(2):219-222. | |
| 18 | 张秀.红景天属植物的药用价值[J].中国野生植物资源,1993(2):33-35. |
| ZHANG X.The medicinal value of Rhodiola plants[J].Chinese Wild Plant Resources,1993(2):33-35. | |
| 19 | TAI T H, TANKSLEY S D.A rapid and inexpensive method for isolation of total DNA from dehydrated plant tissue[J].Plant Molecular Biology Reporter,1990,8(4):297-303. |
| 20 | DING B C, SUN Y, RONG F X,et al.The complete mitochondrial genome of Holothuria spinifera (Théel,1866)[J].Mitochondrial DNA Part B,2020,5(2):1679-1680. |
| 21 | QU X J, MOORE M J, LI D Z,et al.PGA:a software package for rapid,accurate,and flexible batch annotation of plastomes[J].Plant Methods,2019,15(1):50. |
| 22 | ZHENG S, POCZAI P, HYVÖNEN J,et al.Chloroplot:an online program for the versatile plotting of organelle genomes[J].Frontiers in Genetics,2020,11:576124. |
| 23 | SHIELDS D C, SHARP P M.Synonymous codon usage in Bacillus subtilis reflects both translational selection and mutational biases[J].Nucleic Acids Research,1987,15(19):8023-8040. |
| 24 | KATOH K, MISAWA K, KUMA K I,et al.MAFFT:a novel method for rapid multiple sequence alignment based on fast Fourier transform[J].Nucleic Acids Research,2002,30(14):3059-3066. |
| 25 | KALYAANAMOORTHY S, MINH B Q, WONG T K F,et al.ModelFinder:fast model selection for accurate phylogenetic estimates[J].Nature Methods,2017,14(6):587-589. |
| 26 | 刘玉萍,吕婷,朱迪,等.青藏高原特有种—藏扇穗茅叶绿体基因组测序及序列分析[J].植物研究,2018,38(4):518-525. |
| LIU Y P, LÜ T, ZHU D,et al.Sequencing and alignment analysis of the complete chloroplast genome of Littledalea tibetica,an endemic species from the Qinghai-Tibet Plateau[J].Bulletin of Botanical Research,2018,38(4):518-525. | |
| 27 | 兰青阔,陈锐,赵新,等.贝母属药用植物叶绿体基因组单核苷酸多态性位点生物信息学分析[J].食品安全质量检测学报,2018,9(17):4527-4533. |
| LAN Q K, CHEN R, ZHAO X,et al.Bioinformatics analysis for single nucleotide polymorphism sites of chloroplast genome of Fritillaria [J].Journal of Food Safety and Quality,2018,9(17):4527-4533. | |
| 28 | 高雪萍.拟南芥WXR1和WXR3蛋白参与淀粉代谢作用机理的研究[D].泰安:山东农业大学,2015. |
| GAO X P.Functional analysis of the Arabidopsis WXR1,WXR3 proteins during the starch metabolism[D].Tai’an:Shandong Agricultural University,2015. | |
| 29 | CHEN S L, YAO H, HAN J P,et al.Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species[J].PLoS One,2010,5(1):e8613. |
| 30 | 姜汶君,郭梦月,庞晓慧.叶绿体基因组在药用植物鉴定及系统进化研究中的应用[J].世界中医药,2020,15(5):702-708,716. |
| JIANG W J, GUO M Y, PANG X H.Application of chloroplast genome in identification and phylogenetic analysis of medicinal plants[J].World Chinese Medicine,2020,15(5):702-708,716. | |
| 31 | 张慧,何帅兵,孔繁德,等.益母草叶绿体基因组序列与系统进化位置分析[J].中医药信息,2018,35(4):21-27. |
| ZHANG H, HE S B, KONG F D,et al.Sequence of chloroplast genome and the phyletic evolution within Leonurus artemisia [J].Information on Traditional Chinese Medicine,2018,35(4):21-27. | |
| 32 | ZHOU Y X, NIE J, XIAO L,et al.Comparative chloroplast genome analysis of rhubarb botanical origins and the development of specific identification markers[J].Molecules,2018,23(11):2811. |
| 33 | 李素,陈芳,殷缘,等.山新杨PdPapGH12基因克隆及其响应胁迫的组织表达[J].森林工程,2021,37(4):11-21. |
| LI S, CHEN F, YIN Y,et al.Cloning of PdPapGH12 gene of Shanxin poplar (Populus davidiana × P. alba var.pyramidlis) and its tissue expression in response to stress[J].Forest Engineering,2021,37(4):11-21. | |
| 34 | ZHANG T W, FANG Y J, WANG X M,et al.The complete chloroplast and mitochondrial genome sequences of Boea hygrometrica:insights into the evolution of plant organellar genomes[J].PLoS One,2012,7(1):e30531. |
| 35 | DU Q Z, WANG B W, WEI Z Z,et al.Genetic diversity and population structure of Chinese white poplar(Populus tomentosa) revealed by SSR markers[J].Journal of Heredity,2012,103(6):853-862. |
| 36 | 刘凌云.基于转录组测序开发SSR标记解析苔草遗传多样性和种群结构[D].北京:北京林业大学,2020. |
| LIU L Y.Genetic diversity and population structure analysis of Carex L. based on SSR makers generated by RNA-sequencing[D].Beijing:Beijing Forestry University,2020. | |
| 37 | 段义忠,杜忠毓,王海涛.西北干旱区孑遗濒危植物四合木(Tetraena mongolica)叶绿体基因组特征研究及比较分析[J].植物研究,2019,39(5):653-663. |
| DUAN Y Z, DU Z Y, WANG H T.Chloroplast genome characteristics of endangered relict plant Tetraena mongolica in the arid region of northwest China[J].Bulletin of Botanical Research,2019,39 (5):653-663. | |
| 38 | KUANG D Y, WU H, WANG Y L,et al.Complete chloroplast genome sequence of Magnolia kwangsiensis(Magnoliaceae):implication for DNA barcoding and population genetics[J].Genome,2011,54(8):663-673. |
| 39 | HANSEN D R, DASTIDAR S G, CAI Z Q,et al.Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms:Buxus(Buxaceae),Chloranthus(Chloranthaceae),Dioscorea(Dioscoreaceae),and Illicium(Schisandraceae)[J].Molecular Phylogenetics and Evolution,2007,45(2):547-563. |
| 40 | 周晓君,张凯,彭正锋,等.矮牡丹与芍药属其他5个种叶绿体基因组特征的比较[J].林业科学,2020,56(4):82-88. |
| ZHOU X J, ZHANG K, PENG Z F,et al.Comparative analysis of chloroplast genome characteristics between Paeonia jishanensis and other five species of Paeonia [J].Scientia Silvae Sinicae,2020,56(4):82-88. | |
| 41 | KANE N C, CRONK Q.Botany without borders:barcoding in focus[J].Molecular Ecology,2008,17(24):5175-5176. |
| [1] | Xueqin WAN, Yujie SHI, Jinliang HUANG, Jiaxuan MI, Yu ZHONG, Fan ZHANG, Lianghua CHEN. An Overview and Prospect on Taxonomy of the Genus Populus [J]. Bulletin of Botanical Research, 2023, 43(2): 161-168. |
| [2] | Fei WANG, Wenzhi ZHAO, Zhanghong DONG, Luyao MA, Weiying LI, Zongyan LI, Peiyao XIN. Analysis of the Chloroplast Genome Characteristics of 6 Species of Yucca [J]. Bulletin of Botanical Research, 2023, 43(1): 109-119. |
| [3] | Yongchang LU, Xin ZHANG, Luyan ZHANG, Jiuli WANG. Complete Chloroplast Genome Structure and Characterization of Syringa villosa subsp. wolfii [J]. Bulletin of Botanical Research, 2023, 43(1): 120-130. |
| [4] | Yingtong MU, Dongchang FAN, Lijuan LÜ, Xiajie LI, Jingshi LU, Xiaoming ZHANG. Comparison of Codon Characteristics and Phylogeny of Chloroplast Genome between Ranunculaceae and Paeoniaceae [J]. Bulletin of Botanical Research, 2022, 42(6): 964-975. |
| [5] | Mingze XIA, Faqi ZHANG, Xiaofeng CHI, Shuang HAN, Shilong CHEN. Evolutionary Analysis of Chloroplast Genome of Parnassia [J]. Bulletin of Botanical Research, 2022, 42(4): 626-636. |
| [6] | Chen YANG, Xueying YAO, Zhixiang CHEN, Qizhi WANG. Chloroplast Genome Structure and Interspecies Relationship of Sanicula rubriflora [J]. Bulletin of Botanical Research, 2022, 42(3): 437-445. |
| [7] | Dandan SU, Yuping LIU, Tao LIU, Changyuan ZHENG, Yu ZHANG, Yanan WANG, Na QIN, Xu SU. Structure of Chloroplast Genome and its Characteristics of Sphaerophysa salsula [J]. Bulletin of Botanical Research, 2022, 42(3): 446-454. |
| [8] | Qian CAO, Long-Hua XU, Jiu-Li WANG, Fa-Qi ZHANG, Shi-Long CHEN. Molecular Phylogeny of Subtribe Swertiinae [J]. Bulletin of Botanical Research, 2021, 41(3): 408-418. |
| [9] | YANG Bin, MENG Qing-Yao, ZHANG Kai, DUAN Yi-Zhong. Chloroplast Genome Characterization and Identification of Genetic Relationship of Relict Endangered Plant Amygdalus nana [J]. Bulletin of Botanical Research, 2020, 40(5): 686-695. |
| [10] | DUAN Yi-Zhong, DU Zhong-Yu, WANG Hai-Tao. Chloroplast Genome Characteristics of Endangered Relict Plant Tetraena mongolica in the Arid Region of Northwest China [J]. Bulletin of Botanical Research, 2019, 39(5): 653-663. |
| [11] | ZHU Hong, YI Xian-Gui, ZHU Shu-Xia, WANG Hua-Chen, DUAN Yi-Fan, WANG Xian-Rong. Analysis on Relationship and Taxonomic Status of Some Species in Subg. Cerasus Koehne with Chloroplast DNA atpB-rbcL Fragment [J]. Bulletin of Botanical Research, 2018, 38(6): 820-827. |
| [12] | ZHANG Yan-Tong, HUANG Jian, SONG Ju, LIN Li-Mei, FENG Ruo-Xuan, XING Zhao-Bin. Structure and Variation Analysis of Chloroplast Genomes in Fagacea [J]. Bulletin of Botanical Research, 2018, 38(5): 757-765. |
| [13] | WU Po-Po, ZHANG Xue-Jie, ZHANG Luo-Yan, FAN Shou-Jin. Phylogeny and Biogeography Studies of Brachypodieae from China Based on nrITS,psbA-trnH and DMC1 Genes [J]. Bulletin of Botanical Research, 2018, 38(4): 497-505. |
| [14] | LIU Yu-Ping, LÜ Ting, ZHU Di, ZHOU Yong-Hui, LIU Tao, SU Xu. Sequencing and Alignment Analysis of the Complete Chloroplast Genome of Littledalea tibetica,an Endemic Species from the Qinghai-Tibet Plateau [J]. Bulletin of Botanical Research, 2018, 38(4): 518-525. |
| [15] | TANG Li-Li, CHEN Guo-Ping, FENG Xiao-Mei, ZHAO Tie-Jian, SHI Fu-Chen. Community Assembly Rules of the East of Yanshan Mountain Based on Phylogeny [J]. Bulletin of Botanical Research, 2017, 37(6): 807-815. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||