Bulletin of Botanical Research ›› 2022, Vol. 42 ›› Issue (4): 592-601.doi: 10.7525/j.issn.1673-5102.2022.04.009
• Genetic and Breeding • Previous Articles Next Articles
Kun CHEN, Gonggui FANG, Huaizhi MU, Jing JIANG( )
)
Received:2021-11-22
Online:2022-07-20
Published:2022-07-15
Contact:
Jing JIANG
E-mail:jiangjing1960@126.com
About author:CHEN Kun(1995—),male,Ph.D,mainly engaged in tree genetics and breeding research.
Supported by:CLC Number:
Kun CHEN, Gonggui FANG, Huaizhi MU, Jing JIANG. Analysis of the Promoter Sequence and Response Characteristics of the BpPIN3 gene in Betula platyphylla[J]. Bulletin of Botanical Research, 2022, 42(4): 592-601.
Add to citation manager EndNote|Ris|BibTeX
URL: https://bbr.nefu.edu.cn/EN/10.7525/j.issn.1673-5102.2022.04.009
Table 1
Prediction of major cis-acting elements in BpPIN3 promoter region
顺式作用元件 Cis-acting element | 核心序列 Core sequence | 数量 Number | 功能 Function |
|---|---|---|---|
| ABRE | ACGTG | 7 | ABA响应元件 Responsive to ABA |
| ARE | AAACCA | 1 | 厌氧诱导的必须调控元件 Regulatory element essential for anaerobic induction |
| BOX4 | ATTAAT | 1 | 光响应元件 Light responsive element |
| CAAT-box | CAAT | 35 | 顺式作用元件 cis-acting element |
| CCAAT-box | CAACGG | 1 | MYBHv1结合位点 MYBHv1 binding site |
| CGTCA-motif | CGTCA | 1 | MEJA顺式作用调节元件 MEJA cis-acting regulatory element |
| G-box | CACGTT | 7 | 光响应元件 Light responsive element |
| GARE-box | TCTGTTG | 2 | 赤霉素反应元件 Gibberellin-responsive element |
| GT1-motif | GGTTAA | 2 | 光响应元件 Light responsive element |
| HD-Zip 3 | GTAAT(G/C)ATTAC | 1 | 蛋白结合位点 Protein binding site |
| LTR | CCGAAA | 2 | 低温响应顺式作用元件 Low-temperature responsiveness cis-acting element |
| MBS | CAACTG | 1 | MYB结合位点参与干旱诱导MYB binding site involved in drought-inducibility |
| P-box | CCTTTG | 1 | 赤霉素反应元件 Gibberellin-responsive element |
| TATA-box | TATA | 58 | 转录起始-30核心启动子元件 Core promoter element around-30 of transcription start |
| TCA-element | CCATCTTTT | 1 | 水杨酸反应元件 Salicylic-responsive element |
| TCT-motif | TGACG | 1 | 光响应元件Light responsive element |

Fig.3
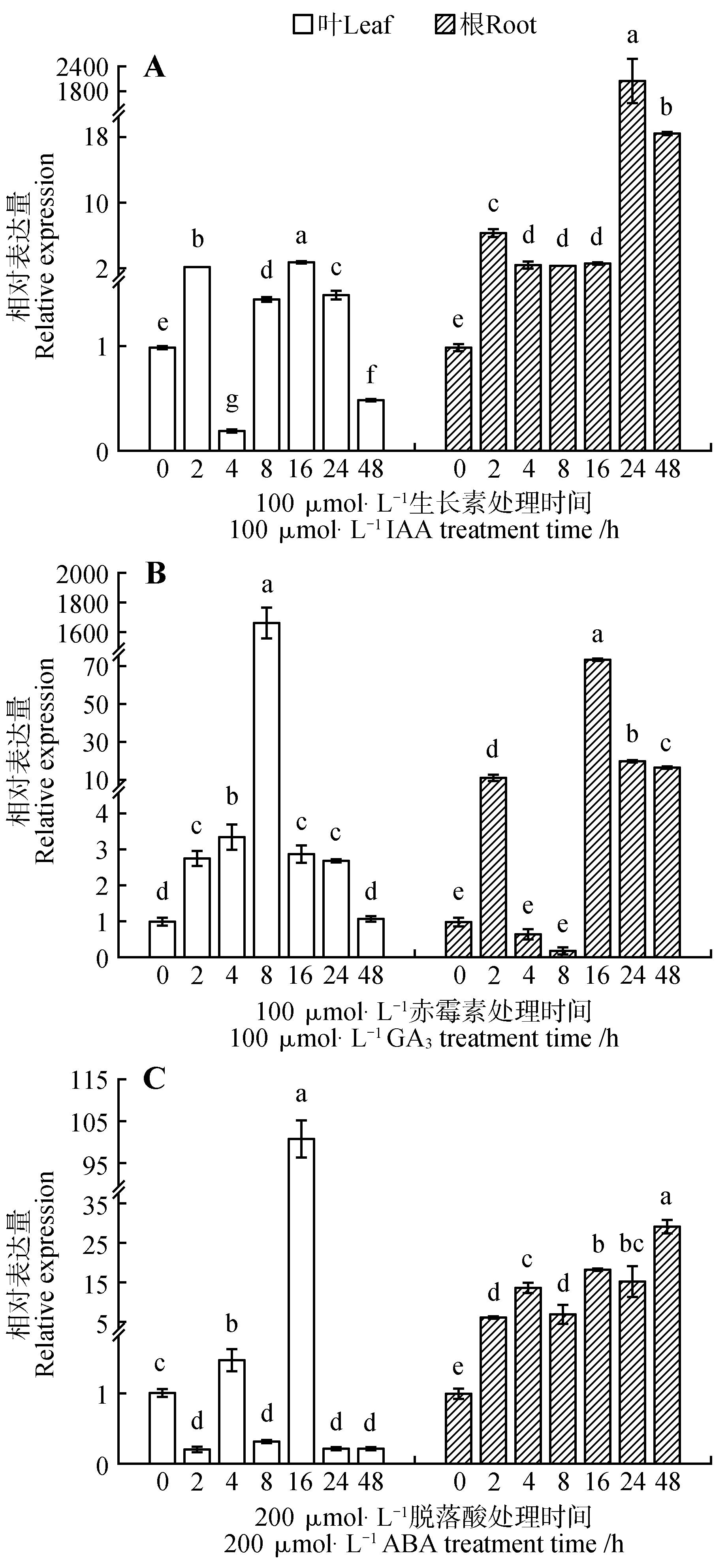
Changes of relative expression of BpPIN3 gene in B. platyphylla after IAA,GA and ABA treatmentA.100 μmol·L-1 IAA treatment;B.100 μmol·L-1 GA3 treatment;C.200 μmol·L-1 ABA treatment;Different lowercase letters represent distinct differences between different treatments(P<0.05);The same as below
| 1 | BOOKER J, CHATFIELD S, LEYSER O.Auxin acts in xylem-associated or medullary cells to mediate apical dominance[J].The Plant Cell,2003,15(2):495-507. |
| 2 | VANNESTE S, FRIML J.Auxin:a trigger for change in plant development[J].Cell,2009,136(6):1005-1016. |
| 3 | BENJAMINS R, SCHERES B.Auxin:the looping star in plant development[J].Annual Review of Plant Biology,2008,59(1):443-465. |
| 4 | PARK J, PARK J, KIM Y,et al.GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis [J].Journal of Biological Chemistry,2007,282(13):10036-10046. |
| 5 | SWARUP R, BENNETT M.Auxin transport:the fountain of life in plants? [J].Development Cell,2003,5(6):824-826. |
| 6 | PETRÁSEK J, MRAVEC J, BOUCHARD R,et al.PIN proteins perform a rate-limiting function in cellular auxin efflux[J].Science,2006,312(5775):914-918. |
| 7 | GRUNEWALD W, FRIML J.The march of the PINs:developmental plasticity by dynamic polar targeting in plant cells[J].The EMBO Journal,2010,29(16):2700-2714. |
| 8 | BLILOU I, XU J, WILDWATER M,et al.The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots[J].Nature,2005,433(7021):39-44. |
| 9 | ALONSO-PERAL M M, CANDELA H, POZO J C DEL,et al.The HVE/CAND1 gene is required for the early patterning of leaf venation in Arabidopsis [J].Development,2006,133(19):3755-3766. |
| 10 | LUSCHNIG C, GAXIOLA R A, GRISAFI P,et al.EIR1,a root-specific protein involved in auxin transport,is required for gravitropism in Arabidopsis thaliana [J].Genes Development,1998,12(14):2175-2187. |
| 11 | CHEN Q, LIU Y, MAERE S,et al.A coherent transcriptional feed-forward motif model for mediating auxin-sensitive PIN3 expression during lateral root development[J].Nature Communication,2015,6:8821. |
| 12 | WILLIGE B C, CHORY J.A current perspective on the role of AGCVIII kinases in PIN-mediated apical hook development[J].Frontiers in Plant Science,2015,6:767. |
| 13 | GANGULY A, LEE S H, CHO M,et al.Differential auxin-transporting activities of PIN-FORMED proteins in Arabidopsis root hair cells[J].Plant Physiology,2010,153(3):1046-1061. |
| 14 | FRIML J, BENKOVÁ E, BLILOU I,et al. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis [J].Cell,2002,108(5):661-673. |
| 15 | DING Z, WANG B, MORENO I,et al.ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis [J].Nature Communications,2012,3(1):941. |
| 16 | MRAVEC J, SKŮPA P, BAILLY A,et al.Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter[J].Nature,2009,459(7250):1136-1140. |
| 17 | KEUSKAMP D H, POLLMANN S, VOESENEK L A C J,et al.Auxin transport through PIN-FORMED3(PIN3) controls shade avoidance and fitness during competition[J].Proceedings of the National Academy of Sciences of the United States of America,2010,107(52):22740-22744. |
| 18 | FRIML J, VIETEN A, SAUER M,et al.Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis [J].Nature,2003,426(6963):147-153. |
| 19 | DING Z, GALVÁN-AMPUDIA C S, DEMARSY E,et al.Light-mediated polarization of the PIN3 auxin transporter for the phototropic response in Arabidopsis [J].Nature Cell Biology,2011,13(4):447-452. |
| 20 | 白羽聪,李翔宇,程占超,等.毛竹PIN基因家族的鉴定与生物信息学分析[J].分子植物育种,2019,17(16):5238-5247. |
| BAI Y C, LI X Y, CHENG Z C,et al.Identification and bioinformatics analysis of PIN gene family in moso bamboo (Phyllostachys edulis)[J].Molecular Plant Breeding,2019,17(16):5238-5247. | |
| 21 | 王占军,杨立伟,徐忠东,等.麻疯树PIN基因家族的鉴定与生物信息学分析[J].分子植物育种,2015,13(5):1111-1122. |
| WANG Z J, YANG L W, XU Z D,et al.Identification and bioinformatics analysis of the PIN gene family in Jatropha curcas [J].Molecular Plant Breeding,2015,13(5):1111-1122. | |
| 22 | VIAENE T, DELWICHE C F, RENSING S A,et al.Origin and evolution of PIN auxin transporters in the green lineage[J].Trends in Plant Science,2013,18(1):5-10. |
| 23 | LIU B B, ZHANG J, WANG L,et al.A survey of Populus PIN-FORMED family genes reveals their diversified expression patterns[J].Journal Experimental Botany,2014,65(9):2437-2448. |
| 24 | BARKOULAS M,HAY A, KOUGIOUMOUTZI E,et al.A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta [J].Nature Genetics,2008,40(9):1136-1141. |
| 25 | FORESTAN C, VAROTTO S.The role of PIN auxin efflux carriers in polar auxin transport and accumulation and their effect on shaping maize development[J].Molecular Plant,2012,5(4):787-798. |
| 26 | SHI Z, JIANG Y, HAN X,et al. SlPIN1 regulates auxin efflux to affect flower abscission process[J].Scientific Reports,2017,7(1):14919. |
| 27 | PATTISON R J, CATALÁ C.Evaluating auxin distribution in tomato(Solanum lycopersicum) through an analysis of the PIN and AUX/LAX gene families[J].The Plant Journal,2012,70(4):585-598. |
| 28 | XIE X D, QIN G Y, SI P,et al.Analysis of Nicotiana tabacum PIN genes identifies NtPIN4 as a key regulator of axillary bud growth[J].Physiologia Plantarum,2017,160(2):222-239. |
| 29 | SHEN C, YUE R, BAI Y,et al.Identification and analysis of Medicago truncatula auxin transporter gene families uncover their roles in responses to Sinorhizobium meliloti infection[J].Plant and Cell Physiology,2015,56(10):1930-1943. |
| 30 | SAŃKO-SAWCZENKO I, ŁOTOCKA B, CZARNOCKA W.Expression analysis of PIN genes in root tips and nodules of Medicago truncatula [J].International Journal of Molecular Sciences,2016,17(8):1197. |
| 31 | ZHANG Y Z, HE P, YANG Z R,et al.A genome-scale analysis of the PIN gene family reveals its functions in cotton fiber development[J].Frontiers Plant Science,2017,8:461. |
| 32 | GOU J Q, STRAUSS S H, TSAI C J,et al.Gibberellins regulate lateral root formation in Populus through interactions with auxin and other hormones[J].The Plant Cell,2010,22(3):623-639. |
| 33 | HAN R, WANG S, LIU C,et al.Transcriptome analysis of a multiple-branches mutant terminal buds in Betula platyphylla × B.pendula [J].Forests,2019,10(5):374. |
| 34 | QU C, BIAN X Y, HAN R,et al.Expression of BpPIN is associated with IAA levels and the formation of lobed leaves in Betula pendula ‘Dalecartica’[J].Journal of Forestry Research,2020,31(1):87-97. |
| 35 | 刘宇,徐焕文,刘桂丰,等.赤霉素GA4+7处理下白桦无性系生长及差异基因表达分析[J].林业科学研究,2017,30(1):181-189. |
| LIU Y, XU H W, LIU G F,et al.Analysis of clonal growth and differences in gene expression of Betula platyphylla × B.pendula under GA4+7 treatment[J].Forest Research,2017,30(1):181-189. | |
| 36 | 黄海娇,李慧玉,姜静. BpAP1转基因白桦中开花相关基因的时序表达[J].东北林业大学学报,2017,45(1):1-6. |
| HUANG H J, LI H Y, JIANG J.Quantitative expression analysis of several flowering-related genes in BpAP1 transgenic birch(Betula platyphylla × Betula pendula)[J].Journal of Northeast Forestry University,2017,45(1):1-6. | |
| 37 | AHUJA I, DE VOS R C H, BONES A M,et al.Plant molecular stress responses face climate change[J].Trends in Plant Science,2010,15(12):664-674. |
| 38 | DU H, LIU H B, XIONG L Z.Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice[J].Frontiers in Plant Science,2013,4:397. |
| 39 | SHANI E, SALEHIN M, ZHANG Y,et al.Plant stress tolerance requires auxin-sensitive Aux/IAA transcriptional repressors[J].Current Biology,2017,27(3):437-444. |
| 40 | ZAHIR Z A.Substrate-dependent auxin production by Rhizobium phaseoli improves the growth and yield of Vigna radiata L.under salt sress conditions[J].Journal of Microbiology and Biotechnology,2010,20(9):1288-1294. |
| 41 | TANAKA H, DHONUKSHE P, BREWER P B,et al.Spatiotemporal asymmetric auxin distribution:a means to coordinate plant development[J].Cellular and Molecular Life Sciences,2006,63(23):2738-2754. |
| 42 | POTTERS G, PASTERNAK T P, GUISEZ Y,et al.Stress-induced morphogenic responses:growing out of trouble?[J].Trends in Plant Science,2007,12(3):98-105. |
| 43 | TOGNETTI V B, AKEN O VAN, MORREEL K,et al.Perturbation of indole-3-butyric acid homeostasis by the UDP-Glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance[J].The Plant Cell,2010,22(8):2660-2679. |
| 44 | CASANOVA-SÁEZ R, VOß U.Auxin metabolism controls developmental decisions in land plants[J].Trends in Plant Science,2019,24(8):741-754. |
| 45 | BRUMOS J, ROBLES L M, YUN J,et al.Local auxin biosynthesis is a key regulator of plant development[J].Developmental Cell,2018,47(3):306-318.e5. |
| 46 | DING Y H, MA Y Z, LIU N,et al.MicroRNAs involved in auxin signalling modulate male sterility under high-temperature stress in cotton(Gossypium hirsutum)[J].The Plant Journal,2017,91(6):977-994. |
| 47 | ZHANG Q, LI J J, ZHANG W J,et al.The putative auxin efflux carrier OsPIN3t is involved in the drought stress response and drought tolerance[J].The Plant Journal,2012,72(5):805-816. |
| 48 | FRIML J.Subcellular trafficking of PIN auxin efflux carriers in auxin transport[J].European Journal of Cell Biology,2010,89(2/3):231-235. |
| 49 | WANG J R, HU H, WANG G H,et al.Expression of PIN genes in rice(Oryza sativa L.):tissue specificity and regulation by hormones[J].Molecular Plant,2009,2(4):823-831. |
| 50 | YUE R,TIE S, SUN T,et al.Genome-wide identification and expression profiling analysis of ZmPIN,ZmPILS,ZmLAX and ZmABCB auxin transporter gene families in maize(Zea mays L.) under various abiotic stresses[J].PLoS One,2015,10(3):e0118751. |
| 51 | 杨蕴力,渠畅,王阳,等.白桦BpPIN5基因启动子组织定位及外源激素应答分析[J].植物研究,2022,42(1):104-111. |
| YANG Y L, QU C, WANG Y,et al.Tissue-specific expression and analysis of exogenous hormone response of BpPIN5 gene promoter in Betula platyphylla [J].Bulletin of Botanical Research,2022,42(1):104-111. | |
| 52 | 王博,曹红利,黄玉婷,等.茶树生长素外运载体基因CsPIN3的克隆与表达分析[J].作物学报,2016,42(1):58-69. |
| WANG B, CAO H L, HUANG Y T,et al.Cloning and expression analysis of auxin efflux carrier gene CsPIN3 in tea plant(Camellia sinensis)[J].Acta Agronomica Sinica,2016,42(1):58-69. | |
| 53 | 李玉军,赵燕,黄丽华,等.十字花科植物PIN3基因调控元件及表达分析[J].中国农业大学学报,2017,22(7):24-33. |
| LI Y J, ZHAO Y, HUANG L H,et al.Regulating elements and expression analysis of PIN3 gene in Brassicaceae family[J].Journal of China Agricultural University,2017,22(7):24-33. | |
| 54 | SEO P J, PARK C M.Auxin homeostasis during lateral root development under drought condition[J].Plant Signaling & Behavior,2009,4(10):1002-1004. |
| 55 | WANG W, DING G D, WHITE P J,et al.Mapping and cloning of quantitative trait loci for phosphorus efficiency in crops:opportunities and challenges[J].Plant and Soil,2019,439(1/2):91-112. |
| 56 | ADAMOWSKI M, FRIML J.PIN-dependent auxin transport:action,regulation,and evolution[J].The Plant Cell,2015,27(1):20-32. |
| 57 | ZHANG X, LIU L, WANG H,et al. MtPIN1 and MtPIN3 play dual roles in regulation of shade avoidance response under different environments in Medicago truncatula [J].International Journal of Molecular Sciences,2020,21(22):8742. |
| [1] | Binghua CHEN, Jie ZHANG, Guifeng LIU, Siting LI, Yuanke GAO, Huiyu LI, Tianfang LI. Selection of Excellent Families and Evaluation of Selection Method for Pulpwood Half-sibling Families of Betula platyphylla [J]. Bulletin of Botanical Research, 2023, 43(5): 690-699. |
| [2] | Jingzhe WANG, Chaokui NIU, Xinyuan LIANG, Chenjing SHEN, Jing YIN. Regulation of Salicylic Acid on Tolerance to Saline Alkali Stress at Seedling Stages of Betula platyphylla [J]. Bulletin of Botanical Research, 2023, 43(3): 379-387. |
| [3] | Jinxia DU, Tingting SHEN, Haoran WANG, Yiping LIN, Huiyu LI, Lianfei ZHANG. Construction of Suppression Expression Vector and Genetic Transformation of BpSPL9 gene from Betula platyphylla [J]. Bulletin of Botanical Research, 2023, 43(1): 30-35. |
| [4] | Xiwu DU, Jun QIN, Kang YE, Yonghong HU, Yiwei TAO, Yongzheng PENG, Yanxiang SHEN, Yan LIANG, Li ZENG. Effects of Flooding Stress on Photosynthetic Characteristics of Yulania stellata and Its Cultivars [J]. Bulletin of Botanical Research, 2022, 42(3): 483-491. |
| [5] | Yunli Yang, Chang Qu, Yang Wang, Guifeng Liu, Jing Jiang. Tissue-specific Expression and Analysis of Exogenous Hormone Response of BpPIN5 Gene Promoter in Betula platyphylla [J]. Bulletin of Botanical Research, 2022, 42(1): 104-111. |
| [6] | Qing MA, Fang-Rui LI, Gui-Feng LIU, Hui-Yu LI. Analysis on the Growth Characters of Betula platyphylla in the Aerospace Mutation [J]. Bulletin of Botanical Research, 2021, 41(4): 540-546. |
| [7] | Yi-Ping GUO, Jia-Xin LIU, Ying YU, Chao WANG, Chuan-Ping YANG. Expression Profile Analysis ofXylem Development Regulated by BpNAC012 Gene from Betula platyphylla [J]. Bulletin of Botanical Research, 2021, 41(2): 251-261. |
| [8] | LIU Jia-Xin, LIU Hui-Zi, SHI Jing-Jing, YU Ying, WANG Chao. Expression of MYB Genes of Birch in Response to Hormones,Salt and Drought [J]. Bulletin of Botanical Research, 2020, 40(5): 743-750. |
| [9] | WANG Wan-Qi, QI Wan-Zhu, ZHAO Qiu-Shuang, ZENG Dong, LIU Yi, FU Peng-Yue, QU Guan-Zheng, ZHAO Xi-Yang. Cloning and Expression Analysis of BpJMJ18 Gene Promoter in Betula platyphylla [J]. Bulletin of Botanical Research, 2020, 40(5): 751-759. |
| [10] | JIANG Cheng, ZHANG Xi, TIAN Qing, LI Li. Isolation of the BpbHLH112 Gene and Expression Analysis of Its Promoter in Betula platyphylla [J]. Bulletin of Botanical Research, 2020, 40(4): 583-592. |
| [11] | SUN Shuo, WANG Xiu-Wei, DU Meng-Tian, LI Jing-Hang, WANG Bo-Yi, LIU Gui-Feng. Root CO2 Efflux Variations of Betula platyphylla Among Sites and Root Diameter Classes in 12 Different Provenances [J]. Bulletin of Botanical Research, 2020, 40(3): 476-480. |
| [12] | QIN Lin-Lin, ZHANG Xi, JIANG Cheng, LI Li. Cloning and Functional Analysis of BpZFP4 Promoter from Birch(Betula platyphylla) [J]. Bulletin of Botanical Research, 2019, 39(6): 917-926. |
| [13] | YAN Bin, WU Dan-Yang, LI Hui-Yu. Genetic Transformation and Resistance Analysis of BpBEE2 Gene from Betula platyphylla [J]. Bulletin of Botanical Research, 2019, 39(2): 287-293. |
| [14] | WANG Sheng-Yu, ZHANG Qi, ZHANG Zheng-Yi, HU Xiao-Qing, TIAN Jing, ZHANG Yong, LIU Xue-Mei. Cloning and Expression Analysis of BpSPL2 Promoter from Betula platyphylla [J]. Bulletin of Botanical Research, 2019, 39(1): 96-103. |
| [15] | WANG Jia-Qi, ZHANG Xi, LI Li. Bioinformatic and Expression Analysis of HD-Zip Family Gene in Betula platyphylla [J]. Bulletin of Botanical Research, 2018, 38(6): 931-938. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||