Bulletin of Botanical Research ›› 2024, Vol. 44 ›› Issue (3): 431-440.doi: 10.7525/j.issn.1673-5102.2024.03.012
• Molecular biology • Previous Articles Next Articles
Lingtong MENG1, Liwei SU1, Xiangxin LI2, Tiansheng XIONG1, Panpeng CHANG1, Mengzhuo LIU1, Chenguang ZHOU1( )
)
Received:2023-11-28
Online:2024-05-20
Published:2024-05-14
Contact:
Chenguang ZHOU
E-mail:zhouchenguang@nefu.edu.cn
CLC Number:
Lingtong MENG, Liwei SU, Xiangxin LI, Tiansheng XIONG, Panpeng CHANG, Mengzhuo LIU, Chenguang ZHOU. Functional Analysis of ANT Transcription Factor of Populus trichocarpa Based on CRISPR-dCas9 Transcription Activation System[J]. Bulletin of Botanical Research, 2024, 44(3): 431-440.
Add to citation manager EndNote|Ris|BibTeX
URL: https://bbr.nefu.edu.cn/EN/10.7525/j.issn.1673-5102.2024.03.012
Table 1
Primer sequences information
引物名称 Primer name | 引物序列(5′→3′) Primer sequences(5′→3′) |
|---|---|
| PtrANT-F | CACCGTGGCTAGACTTGGTCCTGCG |
| PtrANT-R | GCTTCCCTCCACCAATAAGGTCA |
| M13-F | GTAAAACGACGGCCAGT |
| M13-R | GTCATAGCTGTTTCCTG |
| D-ANT-4-F | GTGCTGGTCACCATCACCAA |
| D-ANT-4-R | GGCGTTGATGATCAGGGTGA |
| RT-18s-F | CGAAGACGATCAGATACCGTCCTA |
| RT-18s-R | TTTCTCATAAGGTGCTGGCGGAGGT |
| RT-PtrActin-F | TGTTGCCCTTGACTATGAGCAGGA |
| RT-PtrActin-R | ACGGAATCTCTCAGCTCCAATGGT |
| gRNA1-F | GGTCAAGGGTTCGTTTGTCACACA |
| gRNA1-R | AAACTGTGTGACAAACGAACCCTT |
| gRNA2-F | GGTCAAACACACAAATATTTCTGG |
| gRNA2-R | AAACCCAGAAATATTTGTGTGTTT |
| gRNA3-F | GGTCAGAGACTTCGTTATGTCTGC |
| gRNA3-R | AAACGCAGACATAACGAAGTCTCT |
| TC14-F2 | CAAGCCTGATTGGGAGAAAA |
| zCas9-F12 | GCTGACAAGAAGTACTCGAT |
| Hyg-F | TCCACTATCGGCGAGTACTTC |
| Hyg-R | CAGGACATATCCACGCCCTC |
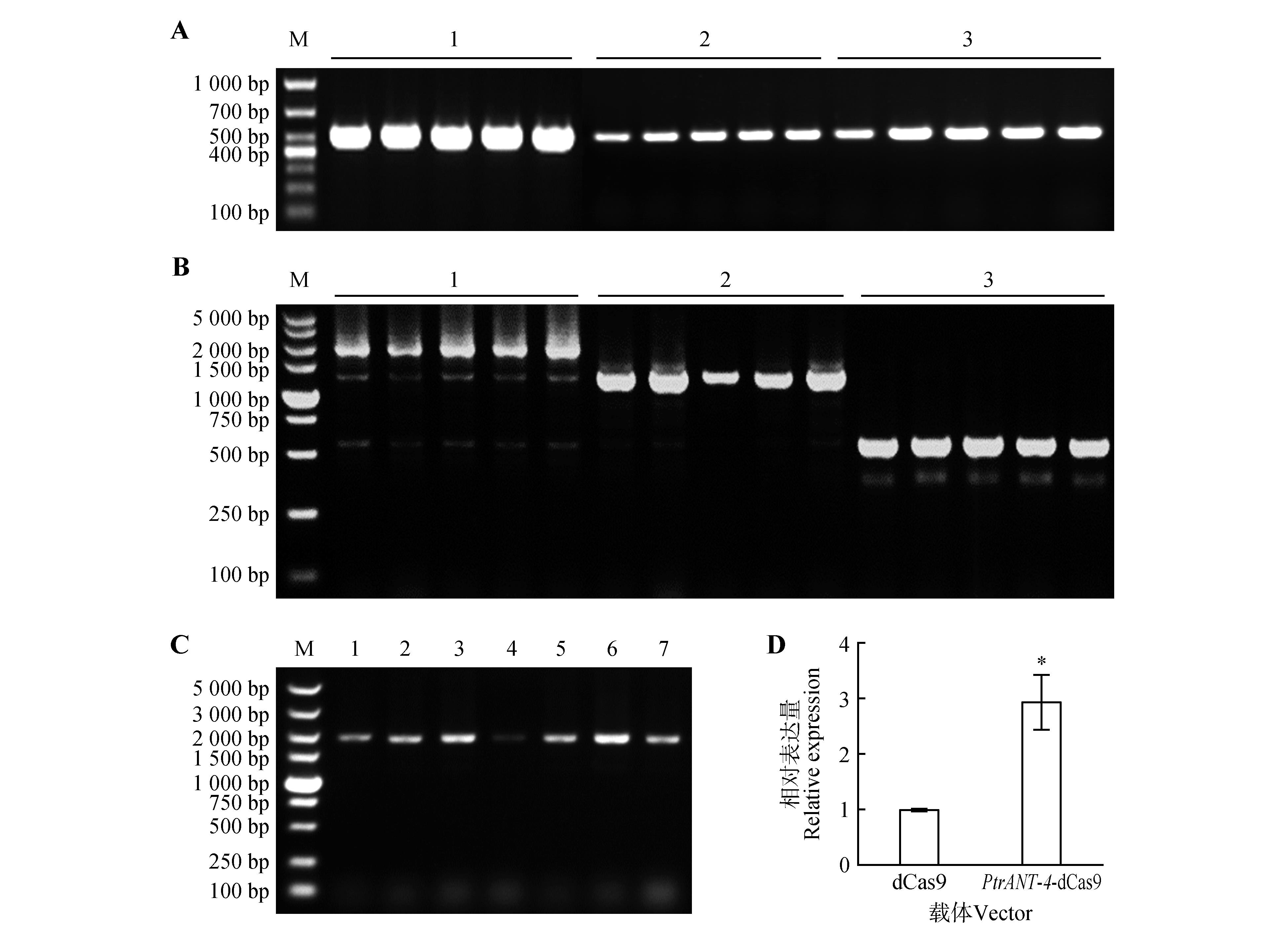
Fig.4
Construction and detection of PtrANT-4 transcription activation vector based on CRISPR-Act3.0 systemA.Construction of entry vectors(1.PCR product of pYPQ131B2.0-gRNA1 colony,2.PCR product of pYPQ132B2.0-gRNA2 colony,3.PCR product of pYPQ133B2.0-gRNA3 colony,M.Marker DL1000);B.Construction of pYPQ143/gRNAs vector(1.PCR product of gRNA1-gRNA3,2.PCR product of gRNA2-gRNA3,3.PCR product of gRNA3,M.Marker DL5000);C.Construction of CRISPR-dCas9/ANTprogRNAs expression vector(1-7.colony PCR products,M.Marker DL5000);D.The transcriptional activation level of PtrANT-4 gene was detected by transient protoplast transformation(error bars represented SE values of three independent experiments,*P<0.05).
| 1 | BORTESI L, FISCHER R.The CRISPR/Cas9 system for plant genome editing and beyond[J].Biotechnology Advances,2015,33(1):41-52. |
| 2 | QI L S, LARSON M H, GILBERT L A,et al.Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression[J].Cell,2013,152(5):1173-1183. |
| 3 | LOWDER L G, ZHANG D W, BALTES N J,et al.A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation[J].Plant Physiology,2015,169(2):971-985. |
| 4 | PIATEK A,ALI Z, BAAZIM H,et al.RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors[J].Plant Biotechnology Journal,2015,13(4):578-589. |
| 5 | LI Z X, ZHANG D D, XIONG X Y,et al.A potent Cas9-derived gene activator for plant and mammalian cells[J].Nature Plants,2017,3(12):930-936. |
| 6 | PAPIKIAN A, LIU W L, GALLEGO-BARTOLOMÉ J,et al.Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems[J].Nature Communications,2019,10(1):729. |
| 7 | LOWDER L G, ZHOU J P, ZHANG Y X,et al.Robust transcriptional activation in plants using multiplexed CRISPR-Act2.0 and mTALE-Act systems[J].Molecular Plant,2018,11(2):245-256. |
| 8 | SELMA S, BERNABÉ-ORTS J M, VAZQUEZ-VILAR M,et al.Strong gene activation in plants with genome-wide specificity using a new orthogonal CRISPR/Cas9-based programmable transcriptional activator[J].Plant Biotechnology Journal,2019,17(9):1703-1705. |
| 9 | PAN C T, WU X C, MARKEL K,et al.CRISPR-Act3.0 for highly efficient multiplexed gene activation in plants[J].Nature Plants,2021,7(7):942-953 |
| 10 | FISCHER U, KUCUKOGLU M, HELARIUTTA Y,et al.The dynamics of cambial stem cell activity[J].Annual Review of Plant Biology,2019,70:293-319. |
| 11 | NIEMINEN K, BLOMSTER T, HELARIUTTA Y,et al.Vascular cambium development[J].The Arabidopsis Book,2015,13:e0177. |
| 12 | LACHAUD S, CATESSON A M, BONNEMAIN J L.Structure and functions of the vascular cambium[J].Comptes Rendus de l'Académie des Sciences - Series III - Sciences de la Vie,1999,322(8):633-650. |
| 13 | EVERT R F.Esau’s Plant anatomy:meristems,cells,and tissues of the plant body:their structure,function,and development[M].New Jersey:John Wiley & Sons,Inc,2006:17. |
| 14 | BERNSTEIN B E, MIKKELSEN T S, XIE X H,et al.A bivalent chromatin structure marks key developmental genes in embryonic stem cells[J].Cell,2006,125(2):315-326. |
| 15 | YOUNG R A.Control of the embryonic stem cell state[J].Cell,2011,144(6):940-954. |
| 16 | HORSTMAN A, WILLEMSEN V, BOUTILIER K,et al.AINTEGUMENTA-LIKE proteins:hubs in a plethora of networks[J].Trends in Plant Science,2014,19(3):146-157. |
| 17 | ELLIOTT R C, BETZNER A S, HUTTNER E,et al.AINTEGUMENTA,an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth[J].The Plant Cell,1996,8(2):155-168. |
| 18 | KRIZEK B A, BLAKLEY I C, HO Y Y,et al.The Arabidopsis transcription factor AINTEGUMENTA orchestrates patterning genes and auxin signaling in the establishment of floral growth and form[J].The Plant Journal,2020,103(2):752-768. |
| 19 | KRIZEK B A, BANTLE A T, HEFLIN J M,et al.AINTEGUMENTA and AINTEGUMENTA-LIKE6 directly regulate floral homeotic,growth,and vascular development genes in young Arabidopsis flowers[J].Journal of Experimental Botany,2021,72(15):5478-5493. |
| 20 | 丁谦,宋晴晴,杨发斌,等.植物ANT类转录因子研究进展[J].农业生物技术学报,2018,26(9):1601-1610. |
| DING Q, SONG Q Q, YANG F B,et al.Research progress of ANT lineage transcription factor in plant[J].Chinese Journal of Agricultural Biotechnology,2018,26(9):1601-1610. | |
| 21 | ZHANG J, ESWARAN G, ALONSO-SERRA J,et al.Transcriptional regulatory framework for vascular cambium development in Arabidopsis roots[J].Nature Plants,2019,5(10):1033-1042. |
| 22 | SCHRADER J, NILSSON J, MELLEROWICZ E,et al.A high-resolution transcript profile across the wood-forming meristem of poplar identifies potential regulators of cambial stem cell identity[J].The Plant Cell,2004,16(9):2278-2292. |
| 23 | ZHANG J, GAO G, CHEN J J,et al.Molecular features of secondary vascular tissue regeneration after bark girdling in Populus [J].New Phytologist,2011,192(4):869-884. |
| 24 | ETCHELLS J P, MISHRA L S, KUMAR M,et al.Wood formation in trees is increased by manipulating PXY-regulated cell division[J].Current Biology,2015,25(8):1050-1055. |
| 25 | DAI X F, ZHAI R, LIN J J,et al.Cell-type-specific PtrWOX4a and PtrVCS2 form a regulatory nexus with a histone modification system for stem cambium development in Populus trichocarpa [J].Nature Plants,2023,9(1):96-111. |
| 26 | LIN Y C, LI W, CHEN H,et al.A simple improved-throughput xylem protoplast system for studying wood formation[J].Nature Protocols,2014,9(9):2194-2205. |
| 27 | LI S J, ZHEN C, XU W J,et al.Simple,rapid and efficient transformation of genotype Nisqually-1:a basic tool for the first sequenced model tree[J].Scientific Reports,2017,7(1):2638. |
| 28 | LI S, LIN Y C J, WANG P Y,et al.The AREB1 transcription factor influences histone acetylation to regulate drought responses and tolerance in Populus trichocarpa [J].The Plant Cell,2019,31(3):663-686. |
| 29 | KRIZEK B A.Ectopic expression of AINTEGUMENTA in Arabidopsis plants results in increased growth of floral organs[J].Developmental Genetics,1999,25(3):224-236. |
| 30 | CHIALVA C, EICHLER E, GRISSI C,et al.Expression of grapevine AINTEGUMENTA-like genes is associated with variation in ovary and berry size[J].Plant Molecular Biology,2016,91(1/2):67-80. |
| 31 | SMETANA O, MÄKILÄ R,LYU M,et al.High levels of auxin signalling define the stem-cell organizer of the vascular cambium[J].Nature,2019,565(7740):485-489. |
| 32 | MÄKILÄ R, WYBOUW B, SMETANA O,et al.Gibberellins promote polar auxin transport to regulate stem cell fate decisions in cambium[J].Nature Plants,2023,9(4):631-644. |
| 33 | GURR S J, RUSHTON P J.Engineering plants with increased disease resistance:what are we going to express?[J].Trends in Biotechnology,2005,23(6):275-282. |
| 34 | ZHENG H Q, LIN S Z, ZHANG Q,et al.Isolation and analysis of a TIR-specific promoter from poplar[J].Forestry Studies in China,2007,9(2):95-106. |
| 35 | 焦勇,柳小庆,江海洋,等.植物组织特异性启动子研究进展[J],中国农业科技导报,2019,21(1):18-28. |
| JIAO Y, LIU X Q, JIANG H Y,et al.Research advances of plant tissue specific promoters[J].Journal of Agricultural Science and Technology,2019,21(1):18-28. | |
| 36 | 张壮,张钱钱,张萌萌,等.植物可诱导基因表达系统的研究进展[J].分子植物育种,2022,20(10):3266-3276. |
| ZHANG Z, ZHANG Q Q, ZHANG M M,et al.Research progress of plant inducible gene expression system[J].Molecular Plant Breeding,2022,20(10):3266-3276. | |
| 37 | WANG H W, ZHANG D H, CHEN M J,et al.Genome editing of 3' UTR-embedded inhibitory region enables generation of gene knock-up alleles in plants[J].Plant Communications,2024,5(3):100745. |
| [1] | Anying HUANG, Dean XIA, Yang ZHANG, Dongchen NA, Qing YAN, Zhigang WEI. Cloning and Drought Tolerance Expression Analysis of PtrWRKY51 Gene in Populus trichocarpa [J]. Bulletin of Botanical Research, 2022, 42(6): 1005-1013. |
| [2] | Xueying WANG, Ruiqi WANG, Yang ZHANG, Cong LIU, Dean XIA, Zhigang WEI. Genome‑wide Identification and Stress Response Analysis of Cyclic Nucleotide-gated Channels(CNGC) Gene Family in Populus trichocarpa [J]. Bulletin of Botanical Research, 2022, 42(4): 613-625. |
| [3] | He CHENG, Shuanghui TIAN, Yang ZHANG, Cong LIU, De’an XIA, Zhigang WEI. Genome-wide Identification and Expression Analysis of nsLTP Gene Family in Populus trichocarpa [J]. Bulletin of Botanical Research, 2022, 42(3): 412-423. |
| [4] | Shuang-Hui TIAN, He CHENG, Yang ZHANG, Cong LIU, De-An XIA, Zhi-Gang WEI. Genome-wide Identification and Expressional Analysis of Carotenoid Cleavage Dioxygenases(CCD) Gene Family in Populus trichocarpa under Drought and Salt Stress [J]. Bulletin of Botanical Research, 2021, 41(6): 993-1005. |
| [5] | Meng-Xuan REN, Yang ZHANG, Shuang WANG, Rui-Qi WANG, Cong LIU, Zhi-Gang WEI. Genome-wide Identification and Expression Analysis GATA Family of Populus trichocarpa [J]. Bulletin of Botanical Research, 2021, 41(1): 107-118. |
| [6] | Bing LI, Yu-Xiang CHENG. Analysis of SHMT Gene Family and Production of PtrSHMT9 Mutant in Populus trichocarpa [J]. Bulletin of Botanical Research, 2020, 40(6): 906-912. |
| [7] | DONG Shi-Wei, YANG Yu-Ning, WANG Nai-Rui, ZHANG Han-Guo, LI Shu-Juan. Gene Cloning and Stress Response Analysis of Natural Disorder Protein in Populus trichocarpa [J]. Bulletin of Botanical Research, 2020, 40(4): 575-582. |
| [8] | LI Ya-Bo, Lü Jia-Xin, TAN Bing, GAO Cai-Qiu. Cloning and expression analysis of 5 ZFP genes from Poplus trichocarpa [J]. Bulletin of Botanical Research, 2020, 40(2): 243-250. |
| [9] | WANG Shuang, ZHANG Yang, REN Meng-Xuan, LIU Ying-Ying, WEI Zhi-Gang. Genome-Wide Analysis of the WOX Family Reveals Their Involvement in Stem Growth of Populus trichocarpa [J]. Bulletin of Botanical Research, 2019, 39(4): 568-577. |
| [10] | WEI Zhen, CHENG Yu-Xiang, XIA De-An. Cas9/gRNA-mediated Targeted PtrFLA31/34 Gene Mutation in Populus [J]. Bulletin of Botanical Research, 2018, 38(5): 741-747. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||