Bulletin of Botanical Research ›› 2021, Vol. 41 ›› Issue (2): 251-261.doi: 10.7525/j.issn.1673-5102.2021.02.013
Previous Articles Next Articles
Yi-Ping GUO, Jia-Xin LIU, Ying YU, Chao WANG, Chuan-Ping YANG( )
)
Received:2020-06-17
Online:2021-03-20
Published:2021-01-05
Contact:
Chuan-Ping YANG
E-mail:yangcp@nefu.edu.cn
About author:GUO Yi-Ping(1994—),female,master,major in molecular mechanism of wood formation.
Supported by:CLC Number:
Yi-Ping GUO, Jia-Xin LIU, Ying YU, Chao WANG, Chuan-Ping YANG. Expression Profile Analysis ofXylem Development Regulated by BpNAC012 Gene from Betula platyphylla[J]. Bulletin of Botanical Research, 2021, 41(2): 251-261.
Add to citation manager EndNote|Ris|BibTeX
URL: https://bbr.nefu.edu.cn/EN/10.7525/j.issn.1673-5102.2021.02.013
Table 1
List of data output quality
样本 Sample | 原始序列 Original sequence | 过滤数据 Filter data | 测序量 Sequencing volume | 错误率 Error rate(%) | Q20(%) | Q30(%) | GC(%) |
|---|---|---|---|---|---|---|---|
| WT_1 | 56242018 | 55322414 | 8.3G | 0.03 | 97.48 | 92.81 | 46.34 |
| WT_2 | 58518124 | 56683910 | 8.5G | 0.03 | 97.55 | 92.97 | 46.30 |
| WT_3 | 52203502 | 50544932 | 7.58G | 0.03 | 97.42 | 92.71 | 46.30 |
| OE_1 | 58644414 | 57292724 | 8.59G | 0.03 | 97.81 | 93.58 | 46.46 |
| OE_2 | 59189588 | 57679272 | 8.65G | 0.03 | 97.34 | 92.52 | 46.55 |
| OE_3 | 56973490 | 56366534 | 8.45G | 0.03 | 97.81 | 93.59 | 46.45 |
| RS_1 | 51023786 | 50389048 | 7.56G | 0.03 | 97.52 | 92.95 | 46.56 |
| RS_2 | 54950226 | 54540828 | 8.18G | 0.03 | 97.33 | 92.47 | 46.51 |
| RS_3 | 57784458 | 57021886 | 8.55G | 0.03 | 97.26 | 92.35 | 46.37 |
Table 3
Gene annotation success rate statistics
基因数量 Number of genes | 百分率 Percentage(%) | |
|---|---|---|
| NR数据库所占比例Proportion of NR database | 117 364 | 65.57 |
| NT数据库所占比例Proportion of NT database | 92 957 | 51.93 |
| KO数据库所占比例Proportion of KO database | 44 707 | 24.97 |
| SwissProt数据库所占比例Proportion of SwissProt database | 86 603 | 48.38 |
| PFAM数据库所占比例Proportion of PFAM database | 84 724 | 47.33 |
| GO数据库所占比例Proportion of GO database | 86 008 | 48.05 |
| KOG数据库所占比例Proportion of KOD database | 29 536 | 16.50 |
| 7大数据库共同出现的比例The proportion of 7 major databases | 18 243 | 10.19 |
| 至少一个数据库注释成功的比例Proportion of at least one database annotation success | 129 213 | 72.19 |
| 总基因数Total genes | 178 983 | 100.00 |
Table 4
Gene Ontology classification of differential gene enrichment in samples
数据库中唯一的标号信息 GO accession gene ontology | 功能的描述信息 Description gene ontology | 为该GO的类别 Term type | 富集分析统计学显著水平 Over represented p-value | 校正后的p-value Corrected p-value | DEG item | DEG list |
|---|---|---|---|---|---|---|
| GO:0001071 | nucleic acid binding transcription factor activity | molecular_function | 5.8636e-17 | 1.7558e-13 | 60 | 581 |
| GO:0003700 | transcription factor activity, sequence-specific DNA binding | molecular_function | 5.8636e-17 | 1.7558e-13 | 60 | 581 |
| GO:0005667 | transcription factor complex | cellular_component | 7.2294e-14 | 1.4432e-10 | 66 | 581 |
| GO:0006355 | regulation of transcription, DNA-templated | biological_process | 1.0952e-08 | 1.3118e-05 | 85 | 581 |

Fig.3
Differential gene GO enrichmentA.Histogram of GO enrichment of differential genes,the abscissa is the GO term of the next three levels of GO, and the ordinate is the number of differential genes annotated under the term(including subterms of the term).The three different classifications represent the three basic classifications of Go term(from left to right are biological processes, cellular components,and molecular functions);B.For GO in Fig.1,according to biological processes,cellular components, and molecular functions,three Histograms of large categories and differential genes up and down classification

Fig.4
KEGG pathway enrichment scatter plotThe vertical axis represents the name of the pathway,and the horizontal axis represents the Rich factor corresponding to the pathway.The size of qvalue is represented by the color of the dot. The smaller the qvalue,the closer the color is to red.To represent.OEvsWT,RSvsWT
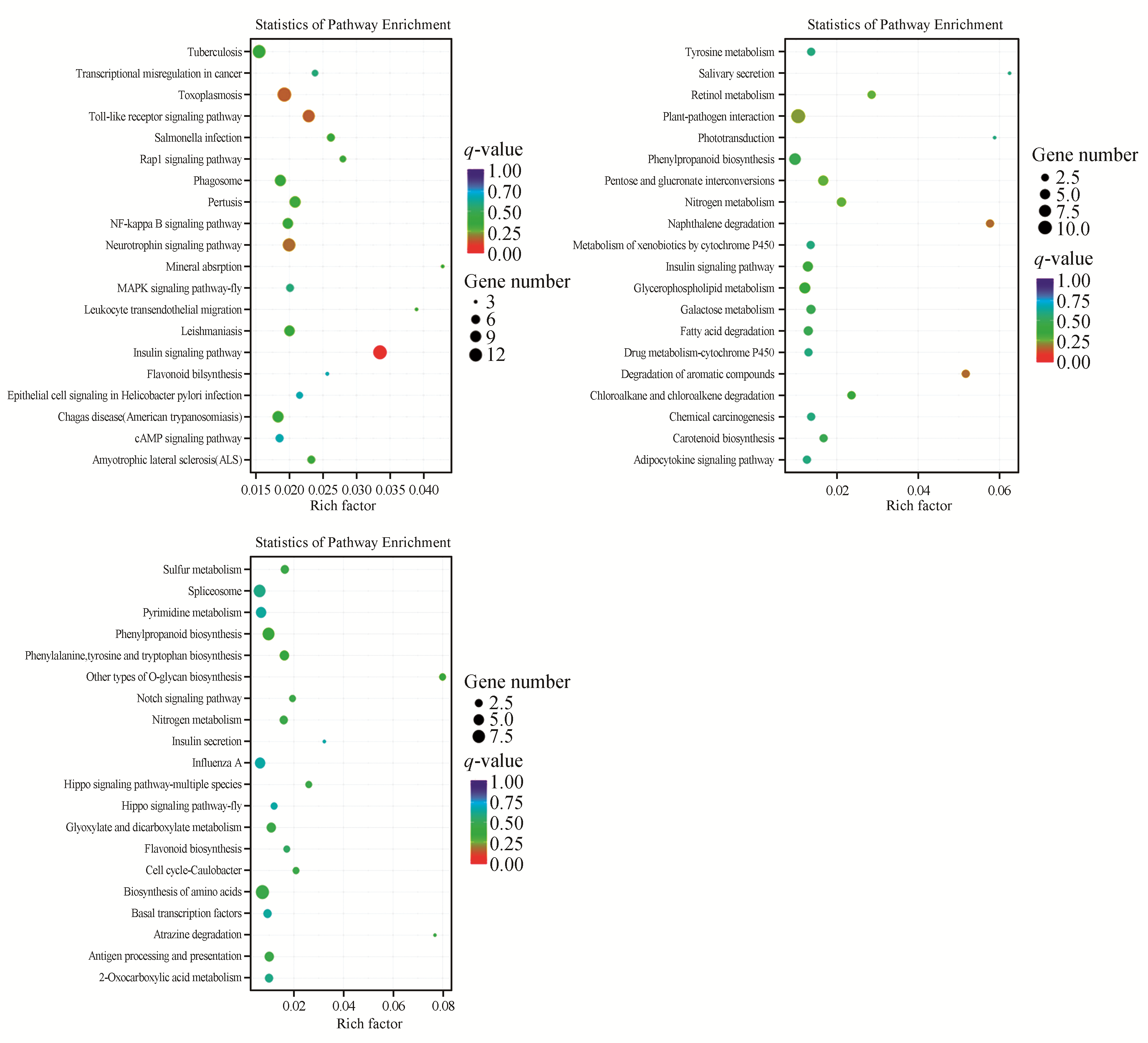
Table 5
Top 10 genes with the highest differential expression fold in overexpressing lines
基因ID Gene_id | log2倍数变化log2 fold change | 基因长度 Gene length | NR GI | NR ID | NR评估 NR evalue | NR注释 NR description |
|---|---|---|---|---|---|---|
| Cluster-12857.66427 | 13.492 | 2 378 | 118132570 | ABK60177.1 | 4.80E-16 | conserved hypothetical protein[Streptomyces ghanaensis ATCC 14672] |
| Cluster-12857.79868 | 11.786 | 4 286 | 645227348 | XP_008220472.1 | 0.00E+00 | PREDICTED: glycine dehydrogenase(decarboxylating),mitochondrial[Prunus mume] |
| Cluster-12857.83191 | 11.500 | 3 565 | 802581682 | XP_012069851.1 | 2.40E-205 | PREDICTED:probable methyltransferase PMT3[Jatropha curcas] |
| Cluster-12857.66430 | 11.254 | 2 133 | 566183478 | XP_002311438.2 | 8.50E-190 | DNA-binding protein RAV2[Populus trichocarpa] |
| Cluster-12857.97128 | 10.632 | 2 699 | 720084019 | XP_010243074.1 | 2.60E-159 | PREDICTED:probable phenylalanine--tRNA ligase alpha subunit[Nelumbo nucifera] |
| Cluster-12857.60814 | 10.592 | 8 563 | 645275167 | XP_008242682.1 | 0.00E+00 | PREDICTED:dnaJ homolog subfamily C GRV2 isoform X2[Prunus mume] |
| Cluster-12857.62522 | 10.323 | 3 186 | 590658081 | XP_007034748.1 | 1.50E-294 | No lysine kinase 1 isoform 1[Theobroma cacao] |
| Cluster-12857.109476 | 10.191 | 1 118 | 590634122 | XP_007028287.1 | 3.30E-116 | 2,4-dienoyl-CoA reductase isoform 2[Theobroma cacao] |
| Cluster-12857.80721 | 9.689 3 | 2 946 | 645260833 | XP_008236004.1 | 1.20E-197 | PREDICTED: amino acid permease 3-like[Prunus mume] |
| Cluster-12857.54251 | 9.565 4 | 945 | 723712802 | XP_010323300.1 | 1.50E-21 | PREDICTED: cucumber peeling cupredoxin-like[Solanum lycopersicum] |
Table 6
The top 10 genes with the highest differential expression folds in the suppressed expression lines
基因ID Gene_id | log2倍数变化 log2 fold change | 基因长度 Gene length | NR GI | NR ID | NR评估 NR evalue | NR说明 NR description |
|---|---|---|---|---|---|---|
| Cluster-12857.25938 | -11.44 8 | 2 910 | 645278673 | XP_008244338.1 | 4.70E-183 | PREDICTED:kinesin-3[Prunus mume] |
| Cluster-12857.85041 | -9.912 4 | 4 383 | 449456208 | XP_004145842.1 | 1.60E-126 | PREDICTED:UTP--glucose-1-phosphate uridylyltransferase[Cucumis sativus] |
| Cluster-12857.98081 | -9.167 2 | 4 075 | 297745750 | CBI15806.3 | 3.60E-35 | unnamed protein product[Vitis vinifera] |
| Cluster-12857.25935 | -9.037 2 | 2 757 | 645278673 | XP_008244338.1 | 4.50E-183 | PREDICTED:kinesin-3[Prunus mume] |
| Cluster-12857.74657 | -8.754 7 | 2 186 | 719976857 | XP_010248668.1 | 1.90E-112 | PREDICTED:uncharacterized protein LOC104591513[Nelumbo nucifera] |
| Cluster-12857.45080 | -8.609 6 | 3 854 | 645230669 | XP_008222040.1 | 2.60E-277 | PREDICTED:ankyrin repeat and sterile alpha motif domain-containing protein 1B-like[Prunus mume] |
| Cluster-12857.58236 | -8.509 5 | 3 080 | 590658081 | XP_007034748.1 | 1.40E-294 | No lysine kinase 1 isoform 1[Theobroma cacao] |
| Cluster-12857.27318 | -8.495 4 | 2 146 | 596174565 | XP_007223177.1 | 5.00E-105 | hypothetical protein PRUPE_ppa009157mg[Prunus persica] |
| Cluster-12857.135896 | -8.463 2 | 1 217 | 590566763 | XP_007010326.1 | 4.60E-132 | Catalytic,putative isoform 1[Theobroma cacao] |
| Cluster-12857.69484 | -8.311 2 | 2 496 | 567877953 | XP_006431535.1 | 1.20E-38 | hypothetical protein CICLE_v10003979mg,partial[Citrus clementina] |
Table 7
Genes related to up-regulated expression of xylem development in overexpressing lines
基因ID Gene_id | log2倍数变化 log2 fold change | 基因长度 Gene length | NR GI | NR ID | NR评估 NR Evalue | NR说明 NR Description |
|---|---|---|---|---|---|---|
| Cluster-12857.118958 | 4.8343 | 1483 | 359497539 | XP_003635559.1 | 1.90E-159 | PREDICTED:cellulose synthase-like protein E6 isoform X1[Vitis vinifera] |
| Cluster-12857.76168 | 2.3144 | 2522 | 225457723 | XP_002277713.1 | 1.70E-274 | PREDICTED: probable cellulose synthase A catalytic subunit 5 [UDP-forming][Vitis vinifera] |
| Cluster-12857.74320 | 3.2946 | 681 | 571534195 | XP_003549490.2 | 5.30E-93 | PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 23-like[Glycine max] |
| Cluster-12857.86571 | 3.5818 | 375 | 590661748 | XP_007035759.1 | 1.10E-60 | Xyloglucan endotransglucosylase/hydrolase family protein [Theobroma cacao] |
| Cluster-12857.94374 | 3.8561 | 1450 | 590683434 | XP_007041599.1 | 7.30E-108 | Fructose-1,6-bisphosphatase, cytosolic[Theobroma cacao] |
| Cluster-12857.94375 | 6.9966 | 1771 | 590683434 | XP_007041599.1 | 7.20E-118 | Fructose-1,6-bisphosphatase, cytosolic[Theobroma cacao] |
| Cluster-12857.76506 | 2.2768 | 2951 | 567877365 | XP_006431272.1 | 1.20E-266 | hypothetical protein CICLE_v10011622mg[Citrus clementina] |
| Cluster-12857.90286 | 5.2815 | 1523 | 645249297 | XP_008230687.1 | 4.90E-155 | PREDICTED:probable galacturonosyltransferase-like 10[Prunus mume] |
| Cluster-12857.59289 | 1.0271 | 7112 | 658001427 | XP_008393176.1 | 9.70E-222 | PREDICTED:galactoside 2-alpha-L-fucosyltransferase-like[Malus domestica] |
| Cluster-12857.85038 | 2.3215 | 3248 | 694386135 | XP_009368869.1 | 5.40E-151 | PREDICTED:glucan endo-1,3-beta-glucosidase 14-like[Pyrus x bretschneideri] |
| Cluster-12857.105697 | 1.9631 | 2202 | 802652705 | XP_012080173.1 | 7.50E-181 | PREDICTED:probable beta-1,3-galactosyltransferase 19[Jatropha curcas] |
| Cluster-12857.77676 | 3.1264 | 1772 | 645247636 | XP_008229928.1 | 3.70E-69 | PREDICTED:fasciclin-like arabinogalactan protein 2[Prunus mume] |
| Cluster-12857.82137 | 8.2031 | 1984 | 166209291 | ABY85195.1 | 2.50E-268 | p-coumaryl-CoA 3'-hydroxylase[Populus alba×Populus grandidentata] |
| Cluster-12857.97128 | 10.632 | 2699 | 720084019 | XP_010243074.1 | 2.60E-159 | PREDICTED:probable phenylalanine—tRNA ligase alpha subunit[Nelumbo nucifera] |
| Cluster-12857.41401 | 4.3742 | 1547 | 854930414 | AKN79297.1 | 8.70E-67 | peroxidase 4[Betula platyphylla] |
| Cluster-12857.76083 | 9.0528 | 1305 | 470108362 | XP_004290489.1 | 5.90E-141 | PREDICTED:peroxidase 12[Fragaria vesca subsp.vesca] |
| Cluster-12857.113235 | 5.9308 | 1124 | 255542656 | XP_002512391.1 | 6.40E-35 | serine-threonine protein kinase,plant-type, putative[Ricinus communis] |
| Cluster-8794.3 | 6.5661 | 2512 | 255555051 | XP_002518563.1 | 3.40E-105 | serine-threonine protein kinase, plant-type, putative[Ricinus communis] |
| Cluster-12857.119083 | 2.6264 | 1800 | 225452312 | XP_002272486.1 | 3.40E-38 | PREDICTED: serine/threonine-protein kinase HT1[Vitis vinifera] |
| Cluster-12857.118054 | 4.7784 | 2738 | 645224059 | XP_008218928.1 | 6.30E-270 | PREDICTED: serine/threonine-protein kinase-like protein CCR4[Prunus mume] |
| Cluster-12857.61436 | 5.542 | 320 | 568883219 | XP_006494380.1 | 6.20E-28 | PREDICTED: wall-associated receptor kinase-like 8-like[Citrus sinensis] |
| Cluster-12857.80854 | 1.1382 | 3198 | 590591190 | XP_007016945.1 | 1.40E-143 | Leucine-rich repeat protein kinase family protein, putative isoform 1[Theobroma cacao] |
| Cluster-12857.46913 | 2.5993 | 1991 | 590640151 | XP_007029875.1 | 2.40E-178 | Leucine-rich repeat containing protein, putative isoform 1[Theobroma cacao] |
Table 8
Genes related to down-regulated expression of xylem development-inhibited expression lines
基因ID gene_id | log2倍数变化 log2 fold change | 基因长度 Gene Length | NR GI | NR ID | NR评估 NR cvalue | NR说明 NR description |
|---|---|---|---|---|---|---|
| Cluster-12857.3401 | -3.592 8 | 2 347 | 590679436 | XP_007040580.1 | 1.10E-299 | Cellulose synthase like G3,putative[Theobroma cacao] |
| Cluster-12857.79817 | -1.296 6 | 3 177 | 541135587 | AGV22123.1 | 3.90E-239 | endo-1,4-beta-glucanase 1[Betula luminifera] |
| Cluster-12857.85041 | -9.912 4 | 4 383 | 449456208 | XP_004145842.1 | 1.60E-126 | PREDICTED:UTP—glucose-1-phosphate uridylyltransferase[Cucumis sativus] |
| Cluster-12857.147630 | -6.796 5 | 494 | 566186337 | XP_002314111.2 | 3.00E-21 | UDP-glucoronosyl/UDP-glucosyl transferase family protein[Populus trichocarpa] |
| Cluster-12857.88974 | -2.119 6 | 1 519 | 702434862 | XP_010069751.1 | 2.30E-165 | PREDICTED:galactinol synthase 2-like[Eucalyptus grandis] |
| Cluster-12857.75094 | -8.027 0 | 5 417 | 470102424 | XP_004287654.1 | 9.40E-92 | PREDICTED: probable polygalacturonase[Fragaria vesca subsp. vesca] |
| Cluster-12857.92483 | -7.776 6 | 5 415 | 470102424 | XP_004287654.1 | 9.40E-92 | PREDICTED:probable polygalacturonase[Fragaria vesca subsp. vesca] |
| Cluster-12857.154648 | -2.077 7 | 837 | 645235470 | XP_008224286.1 | 1.90E-68 | PREDICTED:non-specific lipid-transfer protein-like protein At2g13820[Prunus mume] |
| Cluster-12857.52725 | -7.975 1 | 3 230 | 720011916 | XP_010259705.1 | 7.60E-57 | PREDICTED:probable pectinesterase 29[Nelumbo nucifera] |
| Cluster-12857.57875 | -6.393 4 | 1 137 | 571537497 | XP_003550077.2 | 6.10E-126 | PREDICTED:serine/threonine-protein kinase SRK2I-like isoform 1[Glycine max] |
| Cluster-12857.105258 | -2.206 6 | 1 465 | 645216354 | XP_008220732.1 | 5.60E-100 | PREDICTED:dehydrodolichyl diphosphate synthase 2[Prunus mume] |
| Cluster-12857.122193 | -7.172 0 | 2 231 | 702507890 | XP_010040057.1 | 2.50E-14 | PREDICTED:extensin-like[Eucalyptus grandis] |
| Cluster-12857.20627 | -2.917 3 | 2 070 | 225440560 | XP_002276415.1 | 3.20E-258 | PREDICTED:laccase-7[Vitis vinifera] |
| 1 | Aida M,Ishida T,Fukaki H,et al.Genes involved in organ separation in Arabidopsis:an analysis of the cup-shaped cotyledon mutant[J].Plant Cell,1997,9(6):841-857. |
| 2 | Souer E,Van Houwelingen A,Kloos D,et al.The no apical meristem gene of Petunia is required for pattern formation in embryos and flowers and is expressed at meristem and primordia boundaries[J].Cell,1996,85(2):159-170. |
| 3 | 李少锋.林木木材形成机制及材性改良研究进展[J].温带林业研究,2019,2(2):40-47. |
| Li S F.Wood formation mechanism and properties improvement in forest trees[J].Journal of Temperate Forestry Research,2019,2(2):40-47. | |
| 4 | 姜春妹.杨树油菜素内酯受体基因PtBRI1.2促进木质发育中的功能研究[D].北京:中国科学院大学,2016. |
| Jiang C M.The function of poplar brassinolide receptor gene PtBRI1.2 in promoting lignin development[D].Beijing:University of Chinese Academy of Sciences,2016. | |
| 5 | 李慧,郭晓蕊,刘雅琳,等.木材形成过程中次生壁沉积和细胞程序性死亡的分子调控机制[J].中国科学:生命科学,2020,50(2):123-135. |
| Li H,Guo X R,Liu Y L,et al.The molecular mechanism in secondary wall deposition and programmed cell death of wood formation[J].Scientia Sinica Vitae,2020,50(2):123-135. | |
| 6 | 邓丽萍,任素红,吕建雄,等.树木木质部细胞凋亡及心材形成机理研究进展[J].林产工业,2019,46(6):1-4. |
| Deng L P,Ren S H,Lv J X,et al.Research status on apoptosis in xylem cell of trees and formation mechanism of heartwood[J].China Forest Products Industry,2019,46(6):1-4. | |
| 7 | Mitsuda N,Iwase A,Yamamoto H,et al.NAC transcription factors,NST1 and NST3,are key regulators of the formation of secondary walls in woody tissues of Arabidopsis[J].Plant Cell,2007,19(1):270-280. |
| 8 | 丁忆然.甘蓝型油菜种皮木质素合成相关基因的差异表达分析[D].重庆:西南大学,2019. |
| Ding Y R.Differential expression analysis of lignin synthesis related genes in the seed coat of Brassica napus L.[D].Chongqing:Southwest University,2019. | |
| 9 | Zhong R Q,Lee C,Ye Z H.Functional characterization of poplar wood-associated NAC domain transcription factors[J].Plant Physiology,2010,152(2):1044-1055. |
| 10 | Hu P,Zhang K M,Yang C P.BpNAC012 positively regulates abiotic stress responses and secondary wall biosynthesis[J].Plant Physiology,2019,179(2):700–717. |
| 11 | Wei Z G,Zhang K X,Yang C P,et al.Genetic linkage maps of Betula platyphylla Suk. based on ISSR and AFLP markers[J].Plant Molecular Biology Reporter,2010,28(1):169. |
| 12 | Wang C,Zhang N,Gao C Q,et al.Comprehensive transcriptome analysis of developing xylem responding to artificial bending and gravitational stimuli in Betula platyphylla[J].PLoS One,2014,9(2):e87566. |
| 13 | Schomburg D,Stephan D.UTP-glucose-1-phosphate uridylyltransferase[M].//Schomburg D,Stephan D.Enzyme handbook.Berlin Heidelberg:Springer,1997:163-180. |
| 14 | Sandhoff K,Van Echten G,Schröder M,et al.Metabolism of glycolipids:the role of glycolipid-binding proteins in the function and pathobiochemistry of lysosomes[J].Biochemical Society Transactions,1992,20(3):695-699. |
| 15 | Tanaka K,Murata K,Yamazaki M,et al.Three distinct rice cellulose synthase catalytic subunit genes required for cellulose synthesis in the secondary wall[J].Plant Physiology,2003,133(1):73-83. |
| 16 | Yu L L,Li Q,Zhu Y Y,et al.An auxin-induced β-type endo-1,4-β-glucanase in poplar is involved in cell expansion and lateral root formation[J].Planta,2018,247(5):1149-1161. |
| [1] | Binghua CHEN, Jie ZHANG, Guifeng LIU, Siting LI, Yuanke GAO, Huiyu LI, Tianfang LI. Selection of Excellent Families and Evaluation of Selection Method for Pulpwood Half-sibling Families of Betula platyphylla [J]. Bulletin of Botanical Research, 2023, 43(5): 690-699. |
| [2] | Jingzhe WANG, Chaokui NIU, Xinyuan LIANG, Chenjing SHEN, Jing YIN. Regulation of Salicylic Acid on Tolerance to Saline Alkali Stress at Seedling Stages of Betula platyphylla [J]. Bulletin of Botanical Research, 2023, 43(3): 379-387. |
| [3] | Jinxia DU, Tingting SHEN, Haoran WANG, Yiping LIN, Huiyu LI, Lianfei ZHANG. Construction of Suppression Expression Vector and Genetic Transformation of BpSPL9 gene from Betula platyphylla [J]. Bulletin of Botanical Research, 2023, 43(1): 30-35. |
| [4] | Kun CHEN, Gonggui FANG, Huaizhi MU, Jing JIANG. Analysis of the Promoter Sequence and Response Characteristics of the BpPIN3 gene in Betula platyphylla [J]. Bulletin of Botanical Research, 2022, 42(4): 592-601. |
| [5] | Yunli Yang, Chang Qu, Yang Wang, Guifeng Liu, Jing Jiang. Tissue-specific Expression and Analysis of Exogenous Hormone Response of BpPIN5 Gene Promoter in Betula platyphylla [J]. Bulletin of Botanical Research, 2022, 42(1): 104-111. |
| [6] | Qing MA, Fang-Rui LI, Gui-Feng LIU, Hui-Yu LI. Analysis on the Growth Characters of Betula platyphylla in the Aerospace Mutation [J]. Bulletin of Botanical Research, 2021, 41(4): 540-546. |
| [7] | LIU Jia-Xin, LIU Hui-Zi, SHI Jing-Jing, YU Ying, WANG Chao. Expression of MYB Genes of Birch in Response to Hormones,Salt and Drought [J]. Bulletin of Botanical Research, 2020, 40(5): 743-750. |
| [8] | WANG Wan-Qi, QI Wan-Zhu, ZHAO Qiu-Shuang, ZENG Dong, LIU Yi, FU Peng-Yue, QU Guan-Zheng, ZHAO Xi-Yang. Cloning and Expression Analysis of BpJMJ18 Gene Promoter in Betula platyphylla [J]. Bulletin of Botanical Research, 2020, 40(5): 751-759. |
| [9] | JIANG Cheng, ZHANG Xi, TIAN Qing, LI Li. Isolation of the BpbHLH112 Gene and Expression Analysis of Its Promoter in Betula platyphylla [J]. Bulletin of Botanical Research, 2020, 40(4): 583-592. |
| [10] | SUN Shuo, WANG Xiu-Wei, DU Meng-Tian, LI Jing-Hang, WANG Bo-Yi, LIU Gui-Feng. Root CO2 Efflux Variations of Betula platyphylla Among Sites and Root Diameter Classes in 12 Different Provenances [J]. Bulletin of Botanical Research, 2020, 40(3): 476-480. |
| [11] | QIN Lin-Lin, ZHANG Xi, JIANG Cheng, LI Li. Cloning and Functional Analysis of BpZFP4 Promoter from Birch(Betula platyphylla) [J]. Bulletin of Botanical Research, 2019, 39(6): 917-926. |
| [12] | YAN Bin, WU Dan-Yang, LI Hui-Yu. Genetic Transformation and Resistance Analysis of BpBEE2 Gene from Betula platyphylla [J]. Bulletin of Botanical Research, 2019, 39(2): 287-293. |
| [13] | WANG Sheng-Yu, ZHANG Qi, ZHANG Zheng-Yi, HU Xiao-Qing, TIAN Jing, ZHANG Yong, LIU Xue-Mei. Cloning and Expression Analysis of BpSPL2 Promoter from Betula platyphylla [J]. Bulletin of Botanical Research, 2019, 39(1): 96-103. |
| [14] | WANG Jia-Qi, ZHANG Xi, LI Li. Bioinformatic and Expression Analysis of HD-Zip Family Gene in Betula platyphylla [J]. Bulletin of Botanical Research, 2018, 38(6): 931-938. |
| [15] | CHEN Chen, XING Bao-Yue, BIAN Xiu-Yan, XU Si-Jia, LIU Gui-Feng, JIANG Jing. Growth Characteristics and Endogenous IAA Content of BpCUCt Transgenic Lines in Betula platyphylla [J]. Bulletin of Botanical Research, 2018, 38(4): 506-517. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||