Bulletin of Botanical Research ›› 2023, Vol. 43 ›› Issue (4): 481-492.doi: 10.7525/j.issn.1673-5102.2023.04.001
• Review • Next Articles
Xinyu NI1,2, Junying HE1,2, Mengjiao YAN3, Chao DU1,2( )
)
Received:2022-09-26
Online:2023-07-20
Published:2023-07-03
Contact:
Chao DU
E-mail:duchao@imnu.edu.cn
About author:NI Xinyu(1998—),female,postgraduate,majoring in the research of physiological and molecular biology of plant stress.
Supported by:CLC Number:
Xinyu NI, Junying HE, Mengjiao YAN, Chao DU. Application Progress of RNA-Seq Technology in Rare and Endangered Plants[J]. Bulletin of Botanical Research, 2023, 43(4): 481-492.
Add to citation manager EndNote|Ris|BibTeX
URL: https://bbr.nefu.edu.cn/EN/10.7525/j.issn.1673-5102.2023.04.001
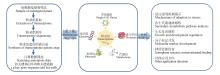
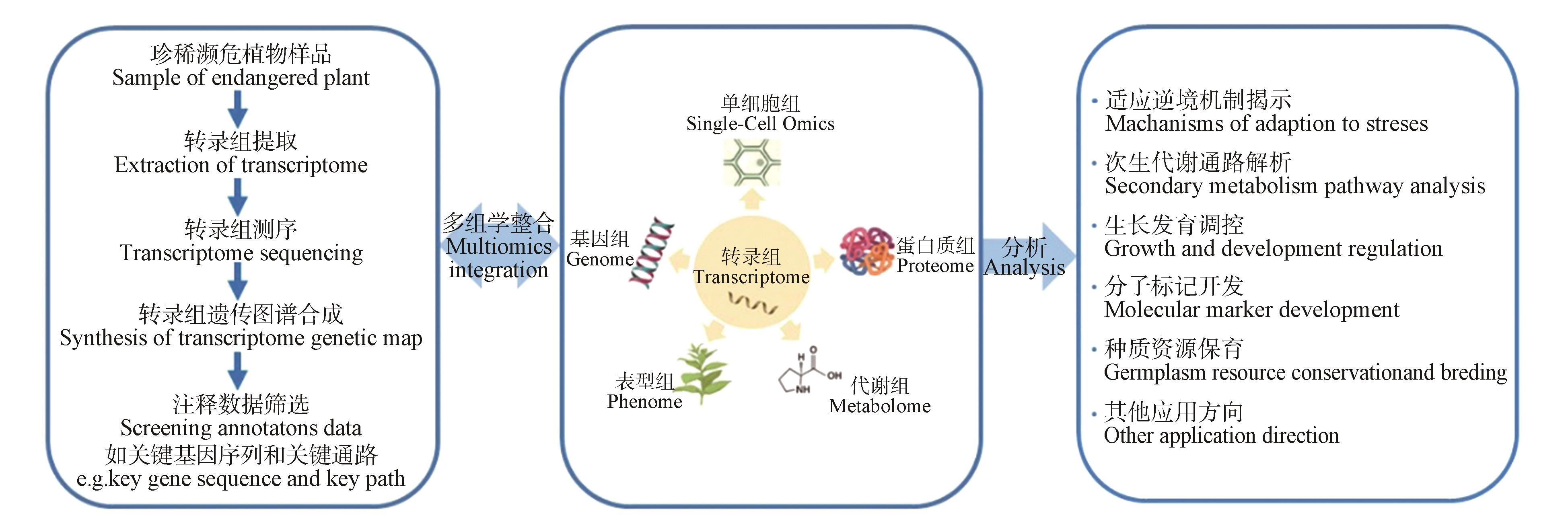
Fig.4
Researchideas on transcriptome of rare and endangered plantsTranscriptome research steps of rare and endangered plants mainly included sample preparation,extraction of transcriptome data,transcriptome sequencing,synthesis of transcriptome genetic map,data screening,etc. In addition,transcriptomics could be the integrated with genome,phenome,proteome,metabolome and single-cell omics to analyze the stress adaptation mechanisms,secondary metabolic pathways,growth and development regulation,development of molecular markers,conservation of germplasm resources of rare and endangered plants
| 1 | 覃海宁,赵莉娜.中国高等植物濒危状况评估[J].生物多样性,2017,25(7):689-695. |
| QIN H N, ZHAO L N.Evaluating the threat status of higher plants in China[J].Biodiversity Science,2017,25(7):689-695. | |
| 2 | 谭聃,欧铜.第三代测序技术的研究进展与临床应用[J].生物工程学报,2022,38(9):3121-3130. |
| TAN D,OU T.Research progress and clinical application of the third-generation sequencing techniques[J].Chinese Journal of Biotechnology,2022,38(9):3121-3130. | |
| 3 | 刘玉洁,胡海洋.第三代测序技术及其在生物学领域的革新[J].科技与创新,2021(5):34-39. |
| LIU Y J, HU H Y.Third-generation sequencing techniques and its innovation in the field of biology[J].Science and Technology & Innovation,2021(5):34-39. | |
| 4 | 刘伟,郭光艳,秘彩莉.转录组学主要研究技术及其应用概述[J].生物学教学,2019,44(10):2-5. |
| LIU W, GUO G Y, BEI C L.A summary of the main research techniques of transcriptology and their applications[J].Biology Teaching,2019,44(10):2-5. | |
| 5 | LOCKHART D J, WINZELER E A.Genomics,gene expression and DNA arrays[J].Nature,2000,405(6788):827-836. |
| 6 | VELCULESCU V E, ZHANG L, ZHOU W,et al.Characterization of the yeast transcriptome[J].Cell,1997,88(2):243-251. |
| 7 | 周华,张新,刘腾云,等.高通量转录组测序的数据分析与基因发掘[J].江西科学,2012,30(5):607-611. |
| ZHOU H, ZHANG X, LIU T Y,et al.Data processing and gene discovery of high-throughput transcriptome sequencing[J].Jiangxi Science,2012,30(5):607-611. | |
| 8 | 张春兰,秦孜娟,王桂芝,等.转录组与RNA-Seq技术[J].生物技术通报,2012(12):51-56. |
| ZHANG C L, QIN Z J, WANG G Z,et al.Transcriptome and RNA-Seq technology[J].Biotechnology Bulletin,2012(12):51-56. | |
| 9 | WANG Z, GERSTEIN M, SNYDER M.RNA-Seq:a revolutionary tool for transcriptomics[J].Nature Reviews Genetics,2009,10(1):57-63. |
| 10 | 杜超.WRKY转录因子家族在植物响应逆境胁迫中的功能及应用[J].草业科学,2021,38(7):1287-1300. |
| DU C.Function and application of the WRKY transcription factor superfamily in plant response to stresses[J].Pratacultural Science,2021,38(7):1287-1300. | |
| 11 | LI L, LI M M, QI X W,et al. De novo transcriptome sequencing and analysis of genes related to salt stress response in Glehnia littoralis [J].PeerJ,2018,6:e5681. |
| 12 | MALA D, AWASTHI S, SHARMA N K,et al.Comparative transcriptome analysis of Rheum australe,an endangered medicinal herb,growing in its natural habitat and those grown in controlled growth chambers[J].Scientific Reports,2021,11(1):3702. |
| 13 | SUN Y B, LIU L H, SUN S K,et al. AnDHN,a dehydrin protein from Ammopiptanthus nanus,mitigates the negative effects of drought stress in plants[J].Frontiers in Plant Science,2021,12:788938. |
| 14 | ZHANG C L, CHEN J H, HUANG W X,et al.Transcriptomics and metabolomics reveal purine and phenylpropanoid metabolism response to drought stress in Dendrobium sinense,an endemic orchid species in Hainan island[J].Frontiers in Genetics,2021,12:692702. |
| 15 | PURAYIL F Y, RAJASHEKAR B, KURUP S S,et al.Transcriptome profiling of Haloxylon persicum (Bunge ex Boiss and Buhse) an endangered plant species under PEG-induced drought stress[J].Genes,2020,11(6):640. |
| 16 | ZHAO D K, SHI Y N, SENTHILKUMAR H A,et al.Enriched networks ‘nucleoside/nucleotide and ribonucleoside/ribonucleotide metabolic processes’ and ‘response to stimulus’ potentially conferred to drought adaptation of the epiphytic orchid Dendrobium wangliangii [J].Physiology and Molecular Biology of Plants,2019,25(1):31-45. |
| 17 | DHIMAN N, SHARMA N K, THAPA P,et al.De novo transcriptome provides insights into the growth behaviour and resveratrol and trans-stilbenes biosynthesis in Dactylorhiza hatagirea:an endangered alpine terrestrial orchid of western Himalaya[J].Scientific Reports,2019,9:13133. |
| 18 | ZHANG J H, ZHU Y J, PAN Y,et al.Transcriptomic profiling and identification of candidate genes in two Phoebe bournei ecotypes with contrasting cold stress responses[J].Trees,2018,32(5):1315-1333. |
| 19 | MENG D L, YU X H, MA L Y,et al.Transcriptomic response of Chinese yew (Taxus chinensis) to cold stress[J].Frontiers in Plant Science,2017,8:468. |
| 20 | POURSALAVATI A, RASHIDI-MONFARED S, EBRAHIMI A.Toward understanding of the methoxylated flavonoid biosynthesis pathway in Dracocephalum kotschyi Boiss[J].Scientific Reports,2021,11(1):19549. |
| 21 | VASHISHT I,PAL T, SOOD H,et al.Comparative transcriptome analysis in different tissues of a medicinal herb,Picrorhiza kurroa pinpoints transcription factors regulating picrosides biosynthesis[J].Molecular Biology Reports,2016,43(12):1395-1409. |
| 22 | DANG Z H, ZHENG L L, WANG J,et al.Transcriptomic profiling of the salt-stress response in the wild recretohalophyte Reaumuria trigyna [J].BMC Genomics,2013,14:29. |
| 23 | BISWAL B, JENA B, GIRI A K,et al. De novo transcriptome and tissue specific expression analysis of genes associated with biosynthesis of secondary metabolites in Operculina turpethum (L.)[J].Scientific Reports,2021,11(1):22539. |
| 24 | WANG Z J, JIANG W M, LIU Y Y,et al.Putative genes in alkaloid biosynthesis identified in Dendrobium officinale by correlating the contents of major bioactive metabolites with genes expression between protocorm-like bodies and leaves[J].BMC Genomics,2021,22(1):579. |
| 25 | SIAH C H, NAMASIVAYAM P, MOHAMED R.Transcriptome reveals senescing callus tissue of Aquilaria malaccensis,an endangered tropical tree,triggers similar response as wounding with respect to terpenoid biosynthesis[J].Tree Genetics & Genomes,2016,12(2):33. |
| 26 | QIAO F, CONG H Q, JIANG X F,et al. De novo characterization of a Cephalotaxus hainanensis transcriptome and genes related to paclitaxel biosynthesis[J].PLoS One,2014,9(9):e106900. |
| 27 | XIANG L, LI Y, ZHU Y J,et al.Transcriptome analysis of the Ophiocordyceps sinensis fruiting body reveals putative genes involved in fruiting body development and cordycepin biosynthesis[J].Genomics,2014,103(1):154-159. |
| 28 | KALRA S, PUNIYA B L, KULSHRESHTHA D,et al. De novo transcriptome sequencing reveals important molecular networks and metabolic pathways of the plant,Chlorophytum borivilianum [J].PLoS One,2013,8(12):e83336. |
| 29 | LIU S S, CHEN J, LI S C,et al.Comparative transcriptome analysis of genes involved in GA-GID1-DELLA regulatory module in symbiotic and asymbiotic seed germination of Anoectochilus roxburghii (Wall.) Lindl.(Orchidaceae)[J].International Journal of Molecular Sciences,2015,16(12):30190-30203. |
| 30 | LI M, DONG X J, PENG J Q,et al. De novo transcriptome sequencing and gene expression analysis reveal potential mechanisms of seed abortion in dove tree(Davidia involucrata Baill.)[J].BMC Plant Biology,2016,16:82. |
| 31 | LIU X T, LIU T F, ZHANG C,et al.Transcriptome profile analysis reveals the regulation mechanism of stamen abortion in Handeliodendron bodinieri [J].Forests,2021,12(8):1071. |
| 32 | TAO L, ZHAO Y, WU Y,et al.Transcriptome profiling and digital gene expression by deep sequencing in early somatic embryogenesis of endangered medicinal Eleutherococcus senticosus Maxim[J].Gene,2016,578(1):17-24. |
| 33 | ZHANG S S, LI Y H, LI Y R,et al.Insights on seed abortion (endosperm and embryo development failure) from the transcriptome analysis of the wild type plant species Paeonia lutea [J].Bioinformation,2020,16(8):638-651. |
| 34 | LI X L, FAN J Z, LUO S M,et al.Comparative transcriptome analysis identified important genes and regulatory pathways for flower color variation in Paphiopedilum hirsutissimum [J].BMC Plant Biology,2021,21(1):495. |
| 35 | 蒋景龙,孙旺,李丽,等.濒危植物秦岭石蝴蝶花瓣数量变异机理研究[J].西北植物学报,2021,41(10):1652-1661. |
| JIANG J L, SUN W, LI L,et al.Study on the variation mechanism of petals number of endangered plant Petrocosmea qinlingensis [J].Acta Botanica Boreali-Occidentalia Sinica,2021,41(10):1652-1661. | |
| 36 | 李美琼,高浦新,朱友林,等.微卫星(SSR)分子标记应用于濒危植物保护的研究进展[J].南方林业科学,2011(2):24-28. |
| LI M Q, GAO P X, ZHU Y L,et al.Research advances in microsatellite (SSR) marker applied in conservation of endangered plants[J].South China Forestry Science,2011(2):24-28. | |
| 37 | XU M, LIU X, WANG J W,et al.Transcriptome sequencing and development of novel genic SSR markers for Dendrobium officinale [J].Molecular Breeding,2017,37(2):18. |
| 38 | LIU H J, TAN W Z, SUN H B,et al.Development and characterization of EST‐SSR markers for Artocarpus hypargyreus (Moraceae)[J].Applications in Plant Sciences,2016,4(12):1600113. |
| 39 | LI C Y, CHIANG T Y, CHIANG Y C,et al.Cross-species,amplifiable EST-SSR markers for Amentotaxus species obtained by next-generation sequencing[J].Molecules,2016,21(1):67. |
| 40 | HU W M.Development of 31 EST-SNP markers in Glycyrrhiza uralensis Fisch (Leguminosae) based on transcriptomics[J].Conservation Genetics Resources,2020,12(2):219-223. |
| 41 | VU D D, SHAH S N M, PHAM M P,et al. De novo assembly and transcriptome characterization of an endemic species of Vietnam,Panax vietnamensis Ha et Grushv.,including the development of EST-SSR markers for population genetics[J].BMC Plant Biology,2020,20(1):358. |
| 42 | XU M, LI Z T, WANG J W,et al.RNA sequencing and SSR marker development for genetic diversity research in Woonyoungia septentrionalis (Magnoliaceae)[J].Conservation Genetics Resources,2018,10(4):867-872. |
| 43 | XU D L, CHEN H B,ACI M,et al. De novo assembly,characterization and development of EST-SSRs from Bletilla striata transcriptomes profiled throughout the whole growing period[J].PLoS One,2018,13(10):e0205954. |
| 44 | ZHOU T, LI Z H, BAI G Q,et al.Transcriptome sequencing and development of genic SSR markers of an endangered Chinese endemic genus Dipteronia oliver (Aceraceae)[J].Molecules,2016,21(3):166. |
| 45 | JIN Y Q, BI Q X, GUAN W B,et al.Development of 23 novel polymorphic EST‐SSR markers for the endangered relict conifer Metasequoia glyptostroboides [J].Applications in Plant Sciences,2015,3(9):1500038. |
| 46 | XIANG X Y, ZHANG Z X, WANG Z G,et al.Transcriptome sequencing and development of EST-SSR markers in Pinus dabeshanensis,an endangered conifer endemic to China[J].Molecular Breeding,2015,35(8):158. |
| 47 | 杨洁.特有濒危植物青檀微卫星(EST-SSR)分子标记开发及其小尺度空间遗传结构研究[D].南京:南京大学,2016. |
| YANG J.Development of polymorphic microsatellite loci and study on fine-scale spatial genetic structure of Pteroceltis tatarinowii,an endangered plant endemic to China[D].Nanjing:Nanjing University,2016. | |
| 48 | 黄蕾.珍稀濒危植物四合木Genic-SSR标记的开发及种群遗传学研究[D].呼和浩特:内蒙古大学,2021. |
| HUANG L.Genic-SSR markers development and population genetic study of the rare and endangered plant Tetraena mongolica [D].Hohhot:Inner Mongolia University,2021. | |
| 49 | ZHOU X J, WANG Y Y, XU Y N,et al. De novo characterization of flower bud transcriptomes and the development of EST-SSR markers for the endangered tree Tapiscia sinensis [J].International Journal of Molecular Sciences,2015,16(6):12855-12870. |
| 50 | HUANG D N, ZHANG Y Q, JIN M D,et al.Characterization and high cross‐species transferability of microsatellite markers from the floral transcriptome of Aspidistra saxicola (Asparagaceae)[J].Molecular Ecology Resources,2014,14(3):569-577. |
| 51 | GUO J D, HUANG Z, SUN J L,et al.Research progress and future development trends in medicinal plant transcriptomics[J].Frontiers in Plant Science,2021,12:691838. |
| 52 | WANG B, REGULSKI M, TSENG E,et al.A comparative transcriptional landscape of maize and sorghum obtained by single-molecule sequencing[J].Genome Research,2018,28(6):921-932. |
| 53 | LI Y P, DAI C, HU C G,et al.Global identification of alternative splicing via comparative analysis of SMRT‐and Illumina‐based RNA‐seq in strawberry[J].The Plant Journal,2017,90(1):164-176. |
| 54 | HOANG N V, FURTADO A, MASON P J,et al.A survey of the complex transcriptome from the highly polyploid sugarcane genome using full-length isoform sequencing and de novo assembly from short read sequencing[J].BMC Genomics,2017,18(1):395. |
| 55 | LI J, HARATA-LEE Y, DENTON M D,et al.Long read reference genome-free reconstruction of a full-length transcriptome from Astragalus membranaceus reveals transcript variants involved in bioactive compound biosynthesis[J].Cell Discovery,2017,3:17031. |
| 56 | 钟雅婷,林艳梅,陈定甲,等.多组学数据整合分析和应用研究综述[J].计算机工程与应用,2021,57(23):1-17. |
| ZHONG Y T, LIN Y M, CHEN D J,et al.Review on integration analysis and application of multi-omics data[J].Computer Engineering and Applications,2021,57(23):1-17. | |
| 57 | LIU Y H, LU S, LIU K F,et al.Proteomics:a powerful tool to study plant responses to biotic stress[J].Plant Methods,2019,15:135. |
| 58 | TRINDADE B M C, REIS R S, VALE E M,et al.Proteomics analysis of the germinating seeds of Cariniana legalis (Mart.) Kuntze (Meliaceae):an endangered species of the Brazilian Atlantic rainforest[J].Brazilian Journal of Botany,2018,41(1):117-128. |
| 59 | PARKASH J, KASHYAP S, KALITA P J,et al.Differential proteomics of Picrorhiza kurrooa Royle ex Benth.in response to dark stress[J].Molecular Biology Reports,2014,41(9):6051-6062. |
| 60 | ZHOU X F, CHEN S L, WU H,et al.Biochemical and proteomics analyses of antioxidant enzymes reveal the potential stress tolerance in Rhododendron chrysanthum Pall[J].Biology Direct,2017,12(1):10. |
| 61 | XIAO Q, MU X L, LIU J S,et al.Plant metabolomics:a new strategy and tool for quality evaluation of Chinese medicinal materials[J].Chinese Medicine,2022,17(1):45. |
| 62 | YU C N, LUO X J, ZHAN X R,et al.Comparative metabolomics reveals the metabolic variations between two endangered Taxus species (T.fuana and T.yunnanensis) in the Himalayas[J].BMC Plant Biology,2018,18(1):197. |
| 63 | ZHENG Y, SUN X, MIAO Y J,et al.A systematic study on the chemical diversity and efficacy of the inflorescence and succulent stem of Cynomorium songaricum [J].Food & Function,2021,12(16):7501-7513. |
| 64 | VERMA R K, IBRAHIM M, FURSULE A,et al.Effect of aging and geographical variations in the content of guggulsterones and metabolomic profiling of oleogum resins of Commiphora wightii:the indian bdellium[J].Pharmacognosy Magazine,2021,17(76):774-779. |
| 65 | AMARAL J, RIBEYRE Z, VIGNEAUD J,et al.Advances and promises of epigenetics for forest trees[J].Forests,2020,11(9):976. |
| 66 | RYU K H, HUANG L, KANG H M,et al.Single-cell RNA sequencing resolves molecular relationships among individual plant cells[J].Plant Physiology,2019,179(4):1444-1456. |
| 67 | SHULSE C N, COLE B J, CIOBANU D,et al.High-throughput single-cell transcriptome profiling of plant cell types[J].Cell Reports,2019,27(7):2241-2247.e4. |
| 68 | LI L B, HUANG G Y, XIANG W B,et al.Integrated transcriptomic and proteomic analyses uncover the regulatory mechanisms of Myricaria laxiflora under flooding stress[J].Frontiers in Plant Science,2022,13:924490. |
| 69 | KAN L, LIAO Q C, CHEN Z P,et al.Dynamic transcriptomic and metabolomic analyses of Madhuca pasquieri (Dubard) H.J.Lam during the post-germination stages[J].Frontiers in Plant Science,2021,12:731203. |
| [1] | Wang Xie, Tianjing Li, Xinyao Li, Fenming Yang, Yin’an Yan, Yongfeng Gao. Identification of PeNAC121 Gene Promoter and Stress Response Pattern Analysis in Populus euphratica [J]. Bulletin of Botanical Research, 2022, 42(2): 234-242. |
| [2] | Yuqi Zhang, Xin Su, Zhiqiang You, Jinbo Fu, Yaguang Zhan, Jing Yin. [J]. Bulletin of Botanical Research, 2022, 42(2): 289-298. |
| [3] | Lian-Bin HAN, Qing GUO, Kai ZHAO, Ting-Bo JIANG, Bo-Ru ZHOU, Li LI. Cloning and Expression Analysis of HD-Zip Transcription Factor PsnHB63 in Populus simonii × P. nigra [J]. Bulletin of Botanical Research, 2021, 41(6): 1006-1014. |
| [4] | Xue-Mei HUANG, Yong-Hong MA, Ting-Fa DONG. Effects of Distance to Riverside on Male and Female Plant Distribution,Plant Morphology and Leaf N-and P-resorption Efficiencies of Cercidiphyllum japonicum [J]. Bulletin of Botanical Research, 2021, 41(5): 789-797. |
| [5] | DONG Shi-Wei, YANG Yu-Ning, WANG Nai-Rui, ZHANG Han-Guo, LI Shu-Juan. Gene Cloning and Stress Response Analysis of Natural Disorder Protein in Populus trichocarpa [J]. Bulletin of Botanical Research, 2020, 40(4): 575-582. |
| [6] | XIAO Zhi-Peng, YIN Chong-Min, GUO Lian-Jin, WU Yuan-Rong, HU Jin-Ping, LIU Yan-Yan, ZHONG You-Chun, XUE Ping-Ping. Effect of Light Quality on Seed Germination and Seeding Growth of Emmenopterys henryi [J]. Bulletin of Botanical Research, 2020, 40(2): 189-195. |
| [7] | GUAN Rui-Ting, LIU Ji-Ming, WANG Min, CHEN Jing-Zhong, WU Meng-Yao, TONG Bing-Li. Genetic Diversity of Critically Endangered Plants Petrocalamus luodianensis from Different Natural Populations [J]. Bulletin of Botanical Research, 2019, 39(5): 740-751. |
| [8] | LI Shuang, XIONG Ying, RALF M;ller-Xing, XING Qian. Distinct Expression Patterns of WRKY6 and PR1 in Arabidopsis Stress Memory Assays [J]. Bulletin of Botanical Research, 2019, 39(5): 752-759. |
| [9] | MA Yue-Hua, GUO Xiao-Rui, YANG Nan, ZHANG Ye, TANG Zhong-Hua, WANG Hong-Zheng. Physiological Mechanisms in Astragalus membranaceus Seedlings Responding to Cadmium Stress [J]. Bulletin of Botanical Research, 2019, 39(4): 497-504. |
| [10] | WANG Meng, ZHNG Dong, LU Yan-Xi, XIAO Hua-Xing, ZHENG Fu-Cong, ZHANG Yu. Multiple Roles of Berberine Bridge Enzyme Gene HbBBE1 in Response to Stress in Hevea brasiliensis Muell. Arg. [J]. Bulletin of Botanical Research, 2018, 38(5): 704-713. |
| [11] | ZHANG Yu, ZHANG Yue, ZHANG Chun-Rui, WANG Yan-Min, WANG Yu-Cheng, WANG Chao. Cloning and Expression Analysis of S-Adenosine Methionine Decarboxylase(ThSAMDC) Gene from Tamarix ramosissima [J]. Bulletin of Botanical Research, 2018, 38(1): 132-140. |
| [12] | ZHU Meng-Yan, YU Bo-Fan, CHEN Hua-Feng. Effects of Nitrate Nitrogen Levels on Physiological Metabolism of Catharanthus roseus [J]. Bulletin of Botanical Research, 2016, 36(4): 535-541. |
| [13] | JIA Yuan-Yuan, ZHANG Chun-Rui, WANG Yu-Cheng, YANG Chuan-Ping, WANG Chao. Cloning and Expression Analysis of a Plasma Membrane Na+/H+ Antiporter Gene in Tamarix hispida [J]. Bulletin of Botanical Research, 2016, 36(3): 380-387. |
| [14] | SONG Xin;DONG Jing-Xiang;LI Kai-Long;LI Hong-Jiao;LI Hui-Yu. Cloning and Identification of a Novel thioredoxin h Gene from Tamarix hisipida [J]. Bulletin of Botanical Research, 2015, 35(3): 340-346. |
| [15] | HE Zhuan-Zhuan;GU Li-Li;LI Xiu-Ming;LAN Hai-Yan*. Expression Analysis and Stress Response Detection of the Phosphoinositide 5-phosphatase gene (CaP5P) in Chenopodium album [J]. Bulletin of Botanical Research, 2013, 33(6): 731-737. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||