Bulletin of Botanical Research ›› 2021, Vol. 41 ›› Issue (5): 729-737.doi: 10.7525/j.issn.1673-5102.2021.05.011
Previous Articles Next Articles
Tian-Tian DU1, Hui-Ping LI1, Bo-Ya WANG1, Qian OU1, Yan HUANG1, Ying CAO1, Shang-Lian HU1,2( )
)
Received:2020-12-29
Online:2021-09-20
Published:2021-07-05
Contact:
Shang-Lian HU
E-mail:hushanglian@126.com;hashanglian@126.com
About author:DU Tian-Tian(1995—),female,master,focused on biochemistry and molecular biology.
Supported by:CLC Number:
Tian-Tian DU, Hui-Ping LI, Bo-Ya WANG, Qian OU, Yan HUANG, Ying CAO, Shang-Lian HU. Cloning and Promotor Analysis of DfMYB3 from Dendrocalamus farinosus[J]. Bulletin of Botanical Research, 2021, 41(5): 729-737.
Add to citation manager EndNote|Ris|BibTeX
URL: https://bbr.nefu.edu.cn/EN/10.7525/j.issn.1673-5102.2021.05.011
Table 1
Primer sequences
引物名称 Primer name | 引物序列(5′→3′) Primer sequences(5′→3′) |
|---|---|
| DfMYB3-F | ATGGGGAGGCATTCTTGTTG |
| DfMYB3-R | CTAGATATGCTCAAAAGATA |
| RT-DfMYB3-F | AAGCAACGGTATGGTATTCGAGCCA |
| RT-DfMYB3-R | GCACATATCCCTTCCATGTTGAATT |
| Tublin-F | GCCGTGAATCTCATCCCCTT |
| Tublin-R | TTGTTCTTGGCATCCCACAT |
| DfMYB3-F(KpnⅠ) | GGGGTACCCCATGGGGAGGCATTCTTGTTG |
| DfMYB3-R(HindⅢ) | CCCAAGCTTGGGGATATGCTCAAAAGATA |
| P3 | TTGCTTGTAGCAACAAGAATGC |
| P2 | TGTTCCCAAGGACAGCATGGAG |
| P1 | TTTCAGGGCGATTCTTATCATA |
| pDfMYB3-F(HindⅢ) | CCAAGCTTGGTATCGAGTTTCGTGTTCCCAAG |
| pDfMYB3-R(XbaI) | GCTCTAGAGCTTCGGTGGAATGCACAACCTGA |

Fig.1
Cloning of DfMYB3 gene from Dendrocalamus farinosus and bioinformatics analysisA.Cloning of DfMYB3 gene from Dendrocalamus farinosus;B.Conserved domain analysis of DfMYB3;C.Secondary structure analysis of DfMYB3(Blue.Alpha helix;Purple.Random coil;Red.Extended strand;Green.Beta turn);D.3D structure analysis of DfMYB3
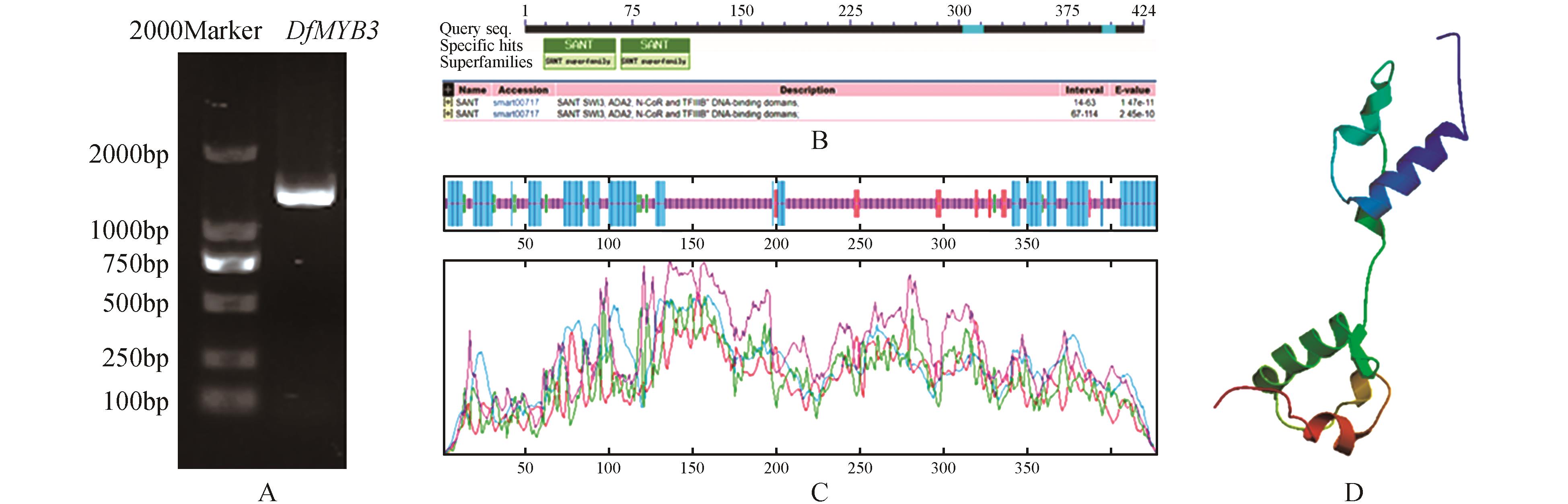
Table 2
Element analysis of DfMYB3 promoter
元件 Site name | 物种 Organism | 位置 Position | 链 Strand | 评分 Matrix score | 序列 Sequence | 功能 Function |
|---|---|---|---|---|---|---|
| ABRE | 拟南芥Arabidopsis thaliana | 705 | + | 5 | ACGTG | cis-acting element involved in the abscisic acid responsiveness |
| ABRE | 大麦Hordeum vulgare | 976 | _ | 9 | CGTACGTGCA | cis-acting element involved in the abscisic acid responsiveness |
| AuxRR-core | 烟草Nicotiana tabacum | 537 | - | 7 | GGTCCAT | cis-acting regulatory element involved in auxin responsiveness |
| Box 4 | 欧芹Petroselinum crispum | 96 | + | 6 | ATTAAT | part of a conserved DNA module involved in light responsiveness |
| CGTCA-motif | 大麦Hordeum vulgare | 191 | + | 5 | CGTCA | cis-acting regulatory element involved in the MeJA-responsiveness |
| CGTCA-motif | 大麦Hordeum vulgare | 813 | - | 5 | CGTCA | cis-acting regulatory element involved in the MeJA-responsiveness |
| CGTCA-motif | 大麦Hordeum vulgare | 604 | - | 5 | CGTCA | cis-acting regulatory element involved in the MeJA-responsiveness |
| CGTCA-motif | 大麦Hordeum vulgare | 854 | - | 5 | CGTCA | cis-acting regulatory element involved in the MeJA-responsiveness |
| G-Box | 豌豆Pisum sativum | 704 | - | 6 | CACGTT | cis-acting regulatory element involved in light responsiveness |
| GARE-motif | 甘草Brassica oleracea | 1017 | - | 7 | TCTGTTG | gibberellin-responsive element |
| GATA-motif | 拟南芥Arabidopsis thaliana | 1574 | - | 7 | GATAGGA | part of a light responsive element |
| GT1-motif | 拟南芥Arabidopsis thaliana | 673 | - | 6 | GGTTAA | light responsive elemen |
| MBS | 拟南芥Arabidopsis thaliana | 841 | + | 6 | CAACTG | MYB binding site involved in drought-inducibility |
| MBS | 拟南芥Arabidopsis thaliana | 1045 | + | 6 | CAACTG | MYB binding site involved in drought-inducibility |
| P-box | 水稻Oryza sativa | 775 | - | 7 | CCTTTTG | gibberellin-responsive element |
| TCA-element | 烟草Nicotiana tabacum | 1108 | - | 9 | CCATCTTTTT | cis-acting element involved in salicylic acid responsiveness |
| TGACG-motif | 大麦Hordeum vulgare | 191 | - | 5 | TGACG | cis-acting regulatory element involved in the MeJA-responsiveness |
| TGACG-motif | Hordeum vulgare | 813 | + | 5 | TGACG | cis-acting regulatory element involved in the MeJA-responsiveness |
| TGACG-motif | Hordeum vulgare | 604 | + | 5 | TGACG | cis-acting regulatory element involved in the MeJA-responsiveness |
| TGACG-motif | Hordeum vulgare | 854 | + | 5 | TGACG | cis-acting regulatory element involved in the MeJA-responsiveness |
| 1 | Pazares J,Ghosal D,Wienand U,et al.The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators [J].Embo Journal,1987,6(12): 3553-3558. |
| 2 | Loguercio L L,Zhang J Q,Wilkins T A.Differential regulation of six novel MYB-domain genes defines two distinct expression patterns in allotetraploid cotton(Gossypium hirsutum L.) [J].Molecular and General Genetics MGG,1999,261(4-5):660-671. |
| 3 | Miyake K,Ito T,Senda M,et al.Isolation of a subfamily of genes for R2R3-MYB transcription factors showing up-regulated expression under nitrogen nutrient-limited conditions[J].Plant Molecular Biology,2003,53(1-2):237-245. |
| 4 | Chen Y H,Yang X Y,He K,et al.The MYB transcription factor superfamily of Arabidopsis:expression analysis and phylogenetic comparison with the rice MYB family[J].Plant Molecular Biology,2006,60(1):107-124. |
| 5 | Stracke R,Werber M,Weisshaar B.The R2R3-MYB gene family in Arabidopsis thaliana[J].Current Opinion in Plant Biology,2001,4(5):447-456. |
| 6 | Suo J F,Liang X,Pu L,et al.Identification of GhMYB109 encoding a R2R3 MYB transcription factor that expressed specifically in fiber initials and elongating fibers of cotton(Gossypium hirsutum L.)[J].Biochimica et Biophysica Acta(BBA)-Gene Structure and Expression,2003,1630(1):25-34. |
| 7 | Kim W C,Kim J Y,Ko J H,et al.Identification of direct targets of transcription factor MYB46 provides insights into the transcriptional regulation of secondary wall biosynthesis[J].Plant Molecular Biology,2014,85(6):589-599. |
| 8 | Uimari A,Strommer J.Myb26:a MYB-like protein of pea flowers with affinity for promoters of phenylpropanoid genes[J].The Plant Journal,1997,12(6):1273-1284. |
| 9 | Ambawat S,Sharma P,Yadav N R,et al.MYB transcription factor genes as regulators for plant responses:an overview[J].Physiology and Molecular Biology of Plants,2013,19(3):307-321. |
| 10 | Huang J F,Chen F,Wu S Y,et al.Cotton GhMYB7 is predominantly expressed in developing fibers and regulates secondary cell wall biosynthesis in transgenic Arabidopsis[J].Science China Life Sciences,2016,59(2):194-205. |
| 11 | Park J S,Kim J B,Cho K J,et al.Arabidopsis R2R3-MYB transcription factor AtMYB60 functions as a transcriptional repressor of anthocyanin biosynthesis in lettuce(Lactuca sativa)[J].Plant Cell Reports,2008,27(6):985-994. |
| 12 | Li H L,Guo D,Peng S Q.Molecular and functional characterization of the JcMYB1,encoding a putative R2R3-MYB transcription factor in Jatropha curcas[J].Plant Growth Regulation,2015,75(1):45-53. |
| 13 | Wang W N,Sun Q,Cai C W,et al.Molecular characterization and alternative splicing of a MYB transcription factor gene in tumourous stem mustard and its response to abiotic stresses[J].Biologia,2016,71(5):538-546. |
| 14 | Fang W B,Ding W W,Zhao X L,et al.Expression profile and function characterization of the MYB type transcription factor genes in wheat(Triticum aestivum L.) under phosphorus deprivation[J].Acta Physiologiae Plantarum,2016,38(1):5. |
| 15 | Yao L M,Jiang Y N,Lu X X,et al.A R2R3-MYB transcription factor from Lablab purpureus induced by drought increases tolerance to abiotic stress in Arabidopsis[J].Molecular Biology Reports,2016,43(10):1089-1100. |
| 16 | Chen Y H,Cao Y Y,Wang L J,et al.Identification of MYB transcription factor genes and their expression during abiotic stresses in maize[J].Biologia Plantarum,2018,62(2):222-230. |
| 17 | 江泽慧.世界竹藤[M].沈阳:辽宁科学技术出版社,2002:367. |
| Jiang Z H.Bamboo and rattan in the world[M].Shenyang:Liaoning Science and Technology Press,2002:367. | |
| 18 | 郭晋隆,李国印,苏亚春,等.甘蔗R2R3-MYB类似基因Sc2RMyb1的克隆及表达特性分析[J].农业生物技术学报,2012,20(9):1009-1017. |
| Guo J L,Li G Y,Su Y C,et al.Cloning and expression analysis of a R2R3-MYB-like gene Sc2RMyb1 from sugarcane(Saccharum complex)[J].Journal of Agricultural Biotechnology,2012,20(9):1009-1017. | |
| 19 | Tian Q Y,Wang X Q,Li C F,et al.Functional characterization of the poplar R2R3-MYB transcription factor PtoMYB216 involved in the regulation of lignin biosynthesis during wood formation[J].PLoS One,2013,8(10):e76369. |
| 20 | Park M Y,Kang J Y,Kim S Y.Overexpression of AtMYB52 confers ABA hypersensitivity and drought tolerance[J].Molecules and Cells,2011,31(5):447-454. |
| 21 | Qin Y X,Wang M C,Tian Y C,et al.Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis[J].Molecular Biology Reports,2012,39(6):7183-7192. |
| 22 | Zhao K M,Bartley L E.Comparative genomic analysis of the R2R3 MYB secondary cell wall regulators of Arabidopsis,poplar,rice,maize,and switchgrass[J].BMC Plant Biology,2014,14:135. |
| 23 | 刘慧春,马广莹,朱开元,等.牡丹PsDREB转录因子基因的克隆及亚细胞定位[J].分子植物育种,2015,13(10):2290-2298. |
| Liu H C,Ma G Y,Zhu K Y,et al.Clonging and subcellular location of a DREB transcription gene of Paeonia suffruticosa[J].Molecular Plant Breeding,2015,13(10):2290-2298. | |
| 24 | Arenhart R A,Schunemann M,Neto L B,et al.Rice ASR1 and ASR5 are complementary transcription factors regulating aluminium responsive genes[J].Plant,Cell & Environment,2016,39(3):645-651. |
| 25 | 张顺仓,冯思念,顾雯,等.丹参中转录因子SmMYB87基因的克隆、亚细胞定位和表达分析[J].中草药,2017,48(17):3597-3604. |
| Zhang S C,Feng S N,Gu W,et al.Cloning,subcellular localization and expression analysis of a transcription factor gene SmMYB87 in Salvia miltiorrhiza[J].Chinese Traditional and Herbal Drugs,2017,48(17):3597-3604. | |
| 26 | 史文君,滕珂,陈露,等.蓝莓VcMYB启动子克隆及其功能初步分析[J].华北农学报,2017,32(2):14-20. |
| Shi W J,Teng K,Chen L,et al.Cloning and functional analysis of VcMYB promoter from Vaccinium corymbosum[J].Acta Agriculturae Boreali-Sinica,2017,32(2):14-20. | |
| 27 | 郭佳茹,胡雪虹,杨郁文,等.海岛棉MYB转录因子GbMYB5基因启动子的克隆及其在拟南芥中的表达特性分析[J].江苏农业学报,2012,28(5):957-962. |
| Guo J R,Hu X H,Yang Y W,et al.Cloning of MYB transcription factor gene(GbMYB5) promoter from Gossypium barbadense L. and functional expression characterization in transgenic Arabidopsis thaliana[J].Jiangsu Journal of Agricultural Sciences,2012,28(5):957-962. | |
| 28 | Prabu G,Theertha Prasad D.Functional characterization of sugarcane MYB transcription factor gene promoter(PScMYBAS1) in response to abiotic stresses and hormones[J].Plant Cell Reports,2012,31(4):661-669. |
| 29 | Xu F C,Liu H L,Xu Y Y,et al.Heterogeneous expression of the cotton R2R3-MYB transcription factor GbMYB60 increases salt sensitivity in transgenic Arabidopsis[J].Plant Cell,Tissue and Organ Culture (PCTOC),2018,133(1):15-25. |
| 30 | 余婧,郭玉双,雷波,等.烟草NtMYB44b转录因子基因克隆以及生物信息学和表达分析[J].植物生理学报,2020,56(2):200-208. |
| Yu J,Guo Y S,Lei B,et al.Cloning and bioinformatic and expression analyses of NtMYB44b transcription factor gene in Nicotiana tabacum[J].Plant Physiology Journal,2020,56(2):200-208. | |
| 31 | Fang Q,Wang Q,Mao H,et al.AtDIV2,an R-R-type MYB transcription factor of Arabidopsis,negatively regulates salt stress by modulating ABA signaling[J].Plant Cell Reports,2018,37(11):1499-1511. |
| 32 | Shan H,Chen S M,Jiang J F,et al.Heterologous expression of the chrysanthemum R2R3-MYB transcription factor CmMYB2 enhances drought and salinity tolerance,increases hypersensitivity to ABA and delays flowering in Arabidopsis thaliana[J].Molecular Biotechnology,2012,51(2):160-173. |
| [1] | ZHU De-Cai, HU Xiao-Qing, ZHAO Tong, TIAN Jing, ZHANG Yong, LIU Xue-Mei. Cloning and Expression Analysis of Promoter of CESA7 Gene in Betula platyphylla [J]. Bulletin of Botanical Research, 2018, 38(4): 551-558. |
| [2] | LI Xi-Yan, ZHAO Kai, ZHANG Xue-Mei, ZHOU Bo-Ru. Expression Analysis of Poplar MYB Transcription Factor Gene Family in Response to Salt Stress [J]. Bulletin of Botanical Research, 2017, 37(3): 424-431. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||